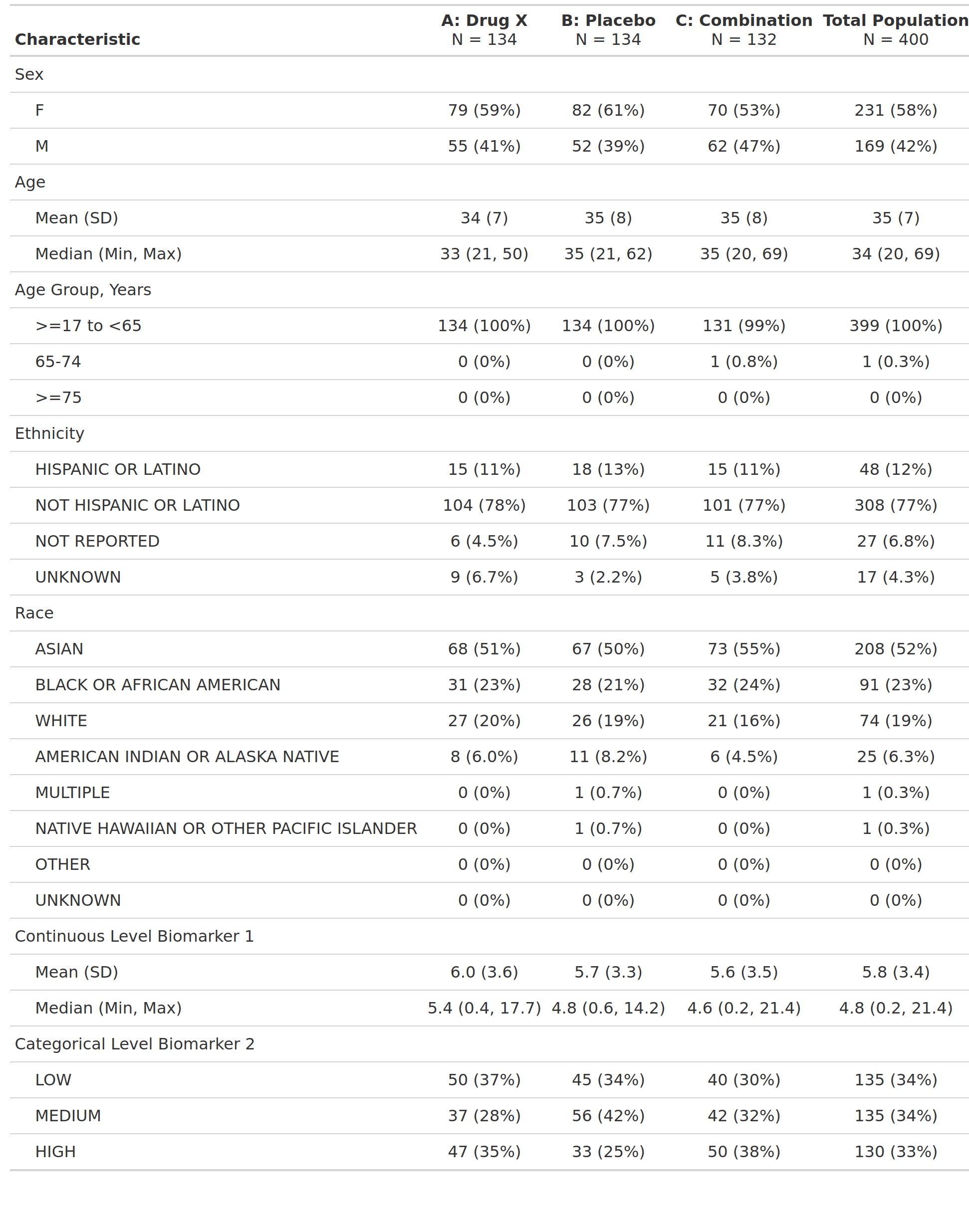
Demographics and Baseline Clinical Characteristics, Safety Population, Pooled Analysis (or Trial X)
FDA Table 02
table
FDA
safety
demographics
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(gtsummary)
adsl <- pharmaverseadam::adsl |>
# removing screen failure observations
filter(TRT01A != "Screen Failure") |>
# Adding a numeric biomarker (weight)
left_join(
pharmaverseadam::advs |>
filter(VSTESTCD == "WEIGHT", VISIT == "BASELINE") |>
select(USUBJID, WEIGHTBL = AVAL),
by = "USUBJID",
relationship = "one-to-one"
)
# Pre-processing --------------------------------------------
# Filter for the safety population, x
data <- adsl |>
filter(SAFFL == "Y")Code
tbl <- data |>
tbl_summary(
by = "TRT01A",
include = c("SEX", "AGE", "AGEGR1", "ETHNIC", "RACE"),
type = all_continuous() ~ "continuous2", # arranges statistics into multiple lines
statistic = list(
all_continuous() ~ c(
"{mean} ({sd})",
"{median} ({min}, {max})"
),
all_categorical() ~ "{n} ({p}%)"
),
label = list(AGEGR1 = "Age Group, Years")
) |>
add_overall(last = TRUE, col_label = "**Total Population** \nN = {N}") |>
# remove default footnote
remove_footnote_header(columns = everything())
tbl
$tbl_summary{cards} data frame: 193 x 12 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 TRT01A Placebo SEX F tabulate n n 53 <fn> stat_1
2 TRT01A Placebo SEX F tabulate N N 86 <fn> stat_1
3 TRT01A Placebo SEX F tabulate p % 0.616 <fn> stat_1
4 TRT01A Placebo SEX M tabulate n n 33 <fn> stat_1
5 TRT01A Placebo SEX M tabulate N N 86 <fn> stat_1
6 TRT01A Placebo SEX M tabulate p % 0.384 <fn> stat_1
7 TRT01A Placebo RACE AMERICAN… tabulate n n 0 <fn> stat_1
8 TRT01A Placebo RACE AMERICAN… tabulate N N 86 <fn> stat_1
9 TRT01A Placebo RACE AMERICAN… tabulate p % 0 <fn> stat_1
10 TRT01A Placebo RACE BLACK OR… tabulate n n 8 <fn> stat_1ℹ 183 more rowsℹ Use `print(n = ...)` to see more rows
$add_overall{cards} data frame: 68 x 10 variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 SEX F tabulate n n 143 <fn> stat_0
2 SEX F tabulate N N 254 <fn> stat_0
3 SEX F tabulate p % 0.563 <fn> stat_0
4 SEX M tabulate n n 111 <fn> stat_0
5 SEX M tabulate N N 254 <fn> stat_0
6 SEX M tabulate p % 0.437 <fn> stat_0
7 RACE AMERICAN… tabulate n n 1 <fn> stat_0
8 RACE AMERICAN… tabulate N N 254 <fn> stat_0
9 RACE AMERICAN… tabulate p % 0.004 <fn> stat_0
10 RACE BLACK OR… tabulate n n 23 <fn> stat_0ℹ 58 more rows
ℹ Use `print(n = ...)` to see more rows