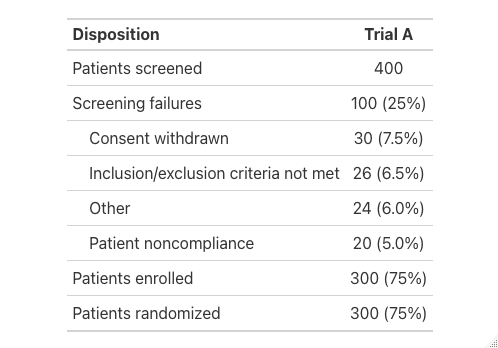
Subject Screening and Enrollment, Screening Population, Trials A and B
FDA Table 03
table
FDA
safety
disposition
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adsl <- pharmaverseadam::adsl
set.seed(1)
scrnfail_reas_lvls <- c(
"Inclusion/exclusion criteria not met", "Subject noncompliance", "Consent withdrawn", "Other"
)
data <- adsl |>
mutate(
ENRLDT = RANDDT,
ENRLFL = !is.na(ENRLDT),
RANDFL = !is.na(RANDDT),
SCRNFL = TRUE,
SCRNFRS = NA_character_
) |>
mutate( # set screen failure reasons for relevant observations.
SCRNFRS = factor(
replace(
SCRNFRS,
TRT01A == "Screen Failure",
sample(scrnfail_reas_lvls, size = sum(TRT01A == "Screen Failure"), replace = TRUE)
),
levels = scrnfail_reas_lvls
)
)Code
tbl_scrn <- tbl_summary(
data = data,
include = "SCRNFL",
missing = "no",
label = ~"Subjects screened"
)
tbl_scrnfrs <- tbl_hierarchical(
data = data,
denominator = adsl,
id = USUBJID,
variables = "SCRNFRS",
label = list(..ard_hierarchical_overall.. = "Screening failures"),
overall_row = TRUE
) |>
modify_indent(columns = label, rows = row_type == "level", indent = 4L) |>
modify_indent(columns = label, rows = variable == "SCRNFRS", indent = 8L)
tbl_enrl_rand <- tbl_summary(
data = data,
include = c("ENRLFL", "RANDFL"),
missing = "no",
label = list(ENRLFL = "Subjects enrolled", RANDFL = "Subjects randomized")
)
tbl <- list(tbl_scrn, tbl_scrnfrs, tbl_enrl_rand) |>
tbl_stack() |>
modify_header(label = "**Disposition**")
tbl
Code
ard <- data |>
ard_stack(
ard_tabulate_value(
variables = SCRNFL,
value = list(SCRNFL = TRUE),
statistic = everything() ~ c("n")
),
ard_tabulate(
variables = SCRNFRS,
statistic = everything() ~ c("n", "p"),
denominator = data
),
ard_tabulate_value(
variables = c(ENRLFL, RANDFL),
value = list(ENRLFL = TRUE, RANDFL = TRUE),
statistic = everything() ~ c("n", "p"),
denominator = data
)
)
ard{cards} data frame: 13 x 9 variable variable_level context stat_name stat_label stat fmt_fun warning error
1 SCRNFL TRUE tabulate… n n 306 0
2 SCRNFRS Inclusio… tabulate n n 17 0
3 SCRNFRS Inclusio… tabulate p % 0.056 <fn>
4 SCRNFRS Subject … tabulate n n 17 0
5 SCRNFRS Subject … tabulate p % 0.056 <fn>
6 SCRNFRS Consent … tabulate n n 11 0
7 SCRNFRS Consent … tabulate p % 0.036 <fn>
8 SCRNFRS Other tabulate n n 7 0
9 SCRNFRS Other tabulate p % 0.023 <fn>
10 ENRLFL TRUE tabulate… n n 254 0
11 ENRLFL TRUE tabulate… p % 0.83 <fn>
12 RANDFL TRUE tabulate… n n 254 0
13 RANDFL TRUE tabulate… p % 0.83 <fn>