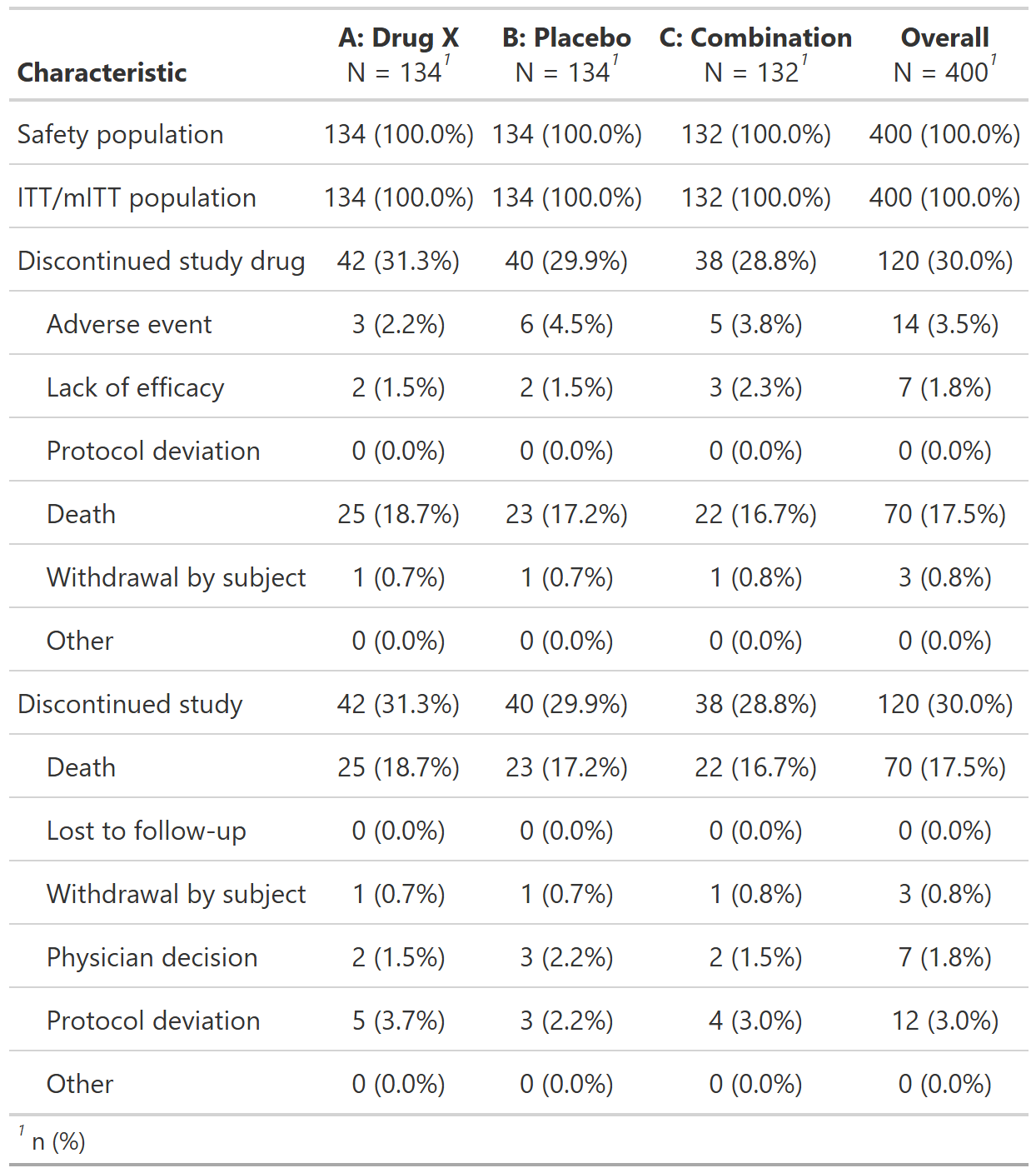
Subject Populations, Randomized Population, Pooled Analysis (or Trial X)
FDA Table 04
table
FDA
safety
disposition
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(gtsummary)
adsl <- pharmaverseadam::adsl |>
filter(TRT01A != "Screen Failure") # Remove screen failure observations
# Pre-processing --------------------------------------------
pop_vars <- c("rand_fl", "ITTFL", "SAFFL", "prot_fl")
lbl_pop_vars <- c("Subjects randomized", "ITT population", "Safety population", "Per-protocol population")
data <- adsl |>
mutate(
across(all_of("SAFFL"), ~ . == "Y"),
ITTFL = !is.na(RANDDT),
rand_fl = !is.na(RANDDT),
prot_fl = EOSSTT == "COMPLETED"
)Code
tbl <- data |>
tbl_summary(
by = "TRT01A",
statistic = list(all_dichotomous() ~ "{n} ({p}%)"),
include = all_of(pop_vars),
digits = list(all_categorical() ~ c(0, 1)),
label = as.list(lbl_pop_vars) |> setNames(pop_vars)
) |>
# Define which variables are children of parent variables
modify_indent(
columns = "label",
rows = variable != "rand_fl",
indent = 4L
) |>
modify_header(label = "**Population**")
tbl
$tbl_summary{cards} data frame: 116 x 12 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 TRT01A Placebo SAFFL TRUE tabulate… n n 86 <fn> stat_1
2 TRT01A Placebo SAFFL TRUE tabulate… N N 86 0 stat_1
3 TRT01A Placebo SAFFL TRUE tabulate… p % 1 <fn> stat_1
4 TRT01A Placebo ITTFL TRUE tabulate… n n 86 <fn> stat_1
5 TRT01A Placebo ITTFL TRUE tabulate… N N 86 0 stat_1
6 TRT01A Placebo ITTFL TRUE tabulate… p % 1 <fn> stat_1
7 TRT01A Placebo rand_fl TRUE tabulate… n n 86 <fn> stat_1
8 TRT01A Placebo rand_fl TRUE tabulate… N N 86 0 stat_1
9 TRT01A Placebo rand_fl TRUE tabulate… p % 1 <fn> stat_1
10 TRT01A Placebo prot_fl TRUE tabulate… n n 58 <fn> stat_1ℹ 106 more rowsℹ Use `print(n = ...)` to see more rows