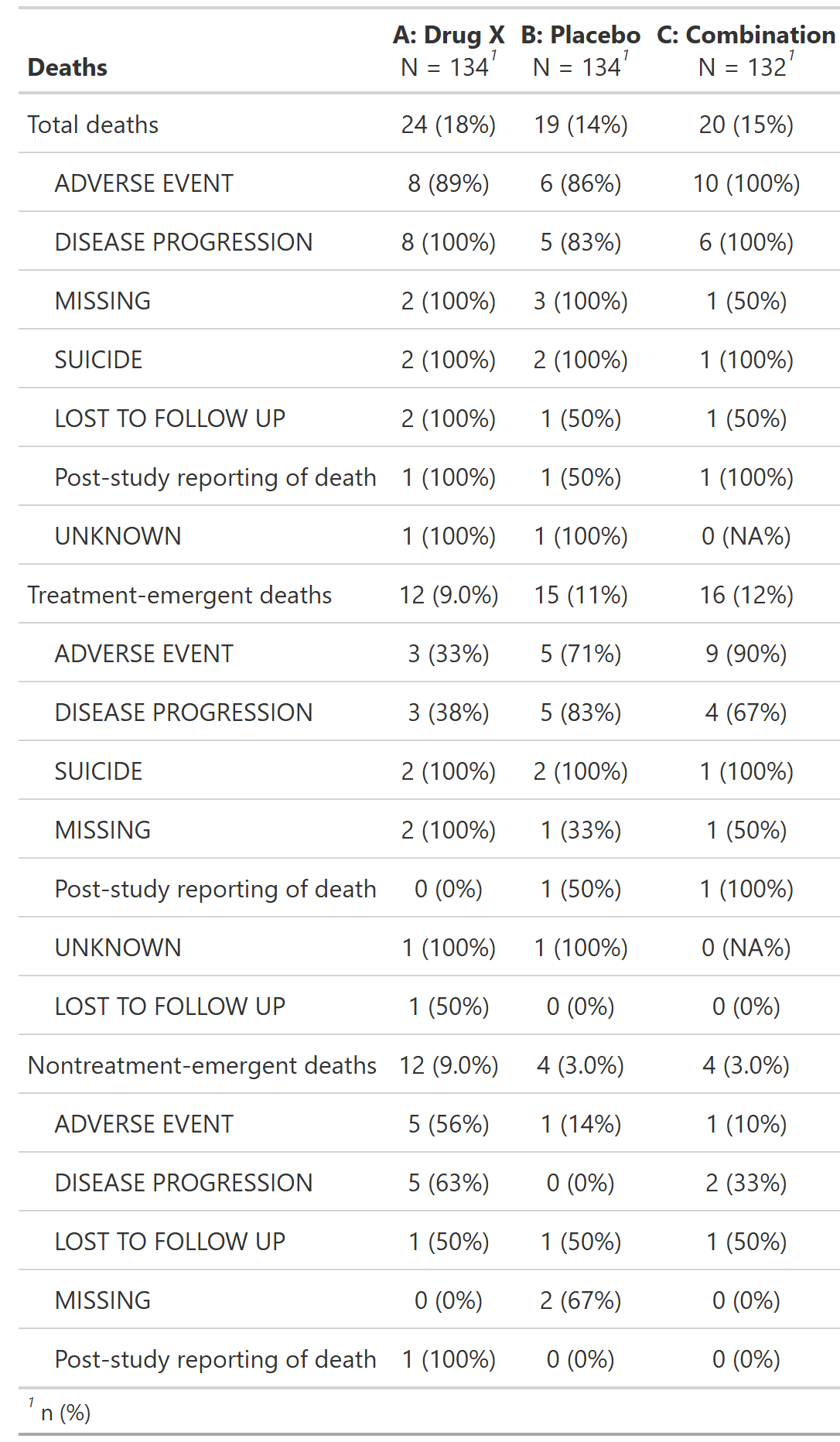
Overview of Adverse Events, Safety Population, Pooled Analysis (or Trial X)
FDA Table 07
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(gtsummary)
library(dplyr)
adsl <- pharmaverseadam::adsl |>
filter(TRT01A != "Screen Failure")
adae <- pharmaverseadam::adae
# Use safety population only
adsl <- adsl |>
filter(SAFFL == "Y")
data <- adae |>
filter(SAFFL == "Y")
set.seed(1)
# this data set doesn't have treatment action recorded. We can generate a variable for demonstration.
aeacn_levels <- c("DRUG INTERRUPTED", "DOSE REDUCED", "DOSE RATE REDUCED", "DOSE INCREASED")
data <- data |>
mutate(AEACN = factor(sample(aeacn_levels, size = nrow(data), replace = TRUE),
levels = aeacn_levels
))Code
# --- PART 1: Overview of serious AEs
# Assign a label to each SAE category
sae_vars <- list(
"SAE" = "AESER",
"Death" = "AESDTH",
"Life-threatening" = "AESLIFE",
"Initial or prolonged hospitalization" = "AESHOSP",
"Disability or permanent damage" = "AESDISAB"
)
# Summarize SAEs per category
tbl_sae <-
lapply(
seq_along(sae_vars),
function(i) { # apply per category
data <- data |>
filter(.data[[sae_vars[[i]]]] == "Y") |>
mutate(!!sae_vars[[i]] := names(sae_vars)[i]) # convert flag to a meaningful value
tbl_hierarchical( # calculate rates
data = data,
variables = all_of(sae_vars[[i]]),
by = TRT01A,
id = USUBJID,
denominator = adsl,
label = sae_vars[[i]] ~ "Event"
)
}
) |>
tbl_stack() |>
modify_indent("label", rows = tbl_id1 != 1, indent = 4L)Column headers among stacked tables differ.
ℹ Use `modify_header()` to update or `quiet = TRUE` to suppress this message.Code
# --- PART 2: Permanent discontinuation
disc_ae <- adae |>
# filtering for adverse events with a "discontinued" End of Study Status
filter(AESER == "Y" & EOSSTT == "DISCONTINUED") |>
mutate(
# convert label to a more meaningful value
AEDISC = "AE leading to permanent discontinuation of treatment"
)
tbl_disc_ae <- tbl_hierarchical( # calculate rates
data = disc_ae,
variables = AEDISC,
by = TRT01A,
id = USUBJID,
denominator = adsl,
label = AEDISC ~ "Event"
)
# -- PART 3: Dose modification
dose_mod_ae <- data |>
mutate(
AEACN = case_when( # convert to meaningful labels
AEACN == "DRUG INTERRUPTED" ~ "AE leading to interruption of study drug",
AEACN == "DOSE REDUCED" ~ "AE leading to reduction of study drug",
AEACN == "DOSE RATE REDUCED" ~ "AE leading to dose delay of study drug",
AEACN == "DOSE INCREASED" ~ "Other",
.default = "Other"
)
)
tbl_dose_mod_ae <- tbl_hierarchical( # calculate rates
data = dose_mod_ae,
denominator = adsl,
id = "USUBJID",
by = "TRT01A",
variables = "AEACN",
label = list(AEACN ~ "Event", ..ard_hierarchical_overall.. = "AE leading to action taken of treatment"),
overall_row = TRUE
) |>
modify_indent(columns = label, rows = variable == "AEACN", indent = 4L)
# -- PART 4: Any AE
tbl_any_ae <- tbl_hierarchical(
data = adae |> mutate(AESEV = factor(.data$AESEV, ordered = TRUE)),
variables = AESEV,
by = TRT01A,
id = USUBJID,
denominator = adsl,
label = list(AESEV ~ "Event", ..ard_hierarchical_overall.. = "Any AE"),
overall_row = TRUE
) |>
modify_indent(columns = label, rows = variable == "AESEV", indent = 4L)
# Put it all together
tbl <- list(tbl_sae, tbl_disc_ae, tbl_dose_mod_ae, tbl_any_ae) |>
tbl_stack() |>
modify_header(label = "**Adverse Event Category**")Column headers among stacked tables differ.
ℹ Use `modify_header()` to update or `quiet = TRUE` to suppress this message.
[[1]]
[[1]][[1]]
[[1]][[1]]$tbl_hierarchical{cards} data frame: 15 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Xanomeli… AESER SAE hierarch… n n 1 1 <fn> stat_1
11 TRT01A Xanomeli… AESER SAE hierarch… N N 72 72 <fn> stat_1
12 TRT01A Xanomeli… AESER SAE hierarch… p % 0.014 1.4 <fn> stat_1
13 TRT01A Xanomeli… AESER SAE hierarch… n n 2 2 <fn> stat_2
14 TRT01A Xanomeli… AESER SAE hierarch… N N 96 96 <fn> stat_2
15 TRT01A Xanomeli… AESER SAE hierarch… p % 0.021 2.1 <fn> stat_2
[[1]][[2]]
[[1]][[2]]$tbl_hierarchical{cards} data frame: 15 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo AESDTH Death hierarch… n n 2 2 <fn> stat_1
11 TRT01A Placebo AESDTH Death hierarch… N N 86 86 <fn> stat_1
12 TRT01A Placebo AESDTH Death hierarch… p % 0.023 2.3 <fn> stat_1
13 TRT01A Xanomeli… AESDTH Death hierarch… n n 1 1 <fn> stat_2
14 TRT01A Xanomeli… AESDTH Death hierarch… N N 96 96 <fn> stat_2
15 TRT01A Xanomeli… AESDTH Death hierarch… p % 0.01 1.0 <fn> stat_2
[[1]][[3]]
[[1]][[3]]$tbl_hierarchical{cards} data frame: 15 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo AESLIFE Life-thr… hierarch… n n 2 2 <fn> stat_1
11 TRT01A Placebo AESLIFE Life-thr… hierarch… N N 86 86 <fn> stat_1
12 TRT01A Placebo AESLIFE Life-thr… hierarch… p % 0.023 2.3 <fn> stat_1
13 TRT01A Xanomeli… AESLIFE Life-thr… hierarch… n n 2 2 <fn> stat_2
14 TRT01A Xanomeli… AESLIFE Life-thr… hierarch… N N 96 96 <fn> stat_2
15 TRT01A Xanomeli… AESLIFE Life-thr… hierarch… p % 0.021 2.1 <fn> stat_2
[[1]][[4]]
[[1]][[4]]$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo AESHOSP Initial … hierarch… n n 5 5 <fn> stat_1
11 TRT01A Placebo AESHOSP Initial … hierarch… N N 86 86 <fn> stat_1
12 TRT01A Placebo AESHOSP Initial … hierarch… p % 0.058 5.8 <fn> stat_1
13 TRT01A Xanomeli… AESHOSP Initial … hierarch… n n 7 7 <fn> stat_2
14 TRT01A Xanomeli… AESHOSP Initial … hierarch… N N 72 72 <fn> stat_2
15 TRT01A Xanomeli… AESHOSP Initial … hierarch… p % 0.097 9.7 <fn> stat_2
16 TRT01A Xanomeli… AESHOSP Initial … hierarch… n n 7 7 <fn> stat_3
17 TRT01A Xanomeli… AESHOSP Initial … hierarch… N N 96 96 <fn> stat_3
18 TRT01A Xanomeli… AESHOSP Initial … hierarch… p % 0.073 7.3 <fn> stat_3
[[1]][[5]]
[[1]][[5]]$tbl_hierarchical{cards} data frame: 12 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Xanomeli… AESDISAB Disabili… hierarch… n n 1 1 <fn> stat_1
11 TRT01A Xanomeli… AESDISAB Disabili… hierarch… N N 96 96 <fn> stat_1
12 TRT01A Xanomeli… AESDISAB Disabili… hierarch… p % 0.01 1.0 <fn> stat_1
[[2]]
[[2]]$tbl_hierarchical{cards} data frame: 15 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Xanomeli… AEDISC AE leadi… hierarch… n n 1 1 <fn> stat_1
11 TRT01A Xanomeli… AEDISC AE leadi… hierarch… N N 72 72 <fn> stat_1
12 TRT01A Xanomeli… AEDISC AE leadi… hierarch… p % 0.014 1.4 <fn> stat_1
13 TRT01A Xanomeli… AEDISC AE leadi… hierarch… n n 2 2 <fn> stat_2
14 TRT01A Xanomeli… AEDISC AE leadi… hierarch… N N 96 96 <fn> stat_2
15 TRT01A Xanomeli… AEDISC AE leadi… hierarch… p % 0.021 2.1 <fn> stat_2
[[3]]
[[3]]$tbl_hierarchical{cards} data frame: 54 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo ..ard_hierarchical_overall.. TRUE hierarch… n n 69 69 <fn> stat_1ℹ 44 more rowsℹ Use `print(n = ...)` to see more rows
[[4]]
[[4]]$tbl_hierarchical{cards} data frame: 45 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo ..ard_hierarchical_overall.. TRUE hierarch… n n 69 69 <fn> stat_1ℹ 35 more rows
ℹ Use `print(n = ...)` to see more rows