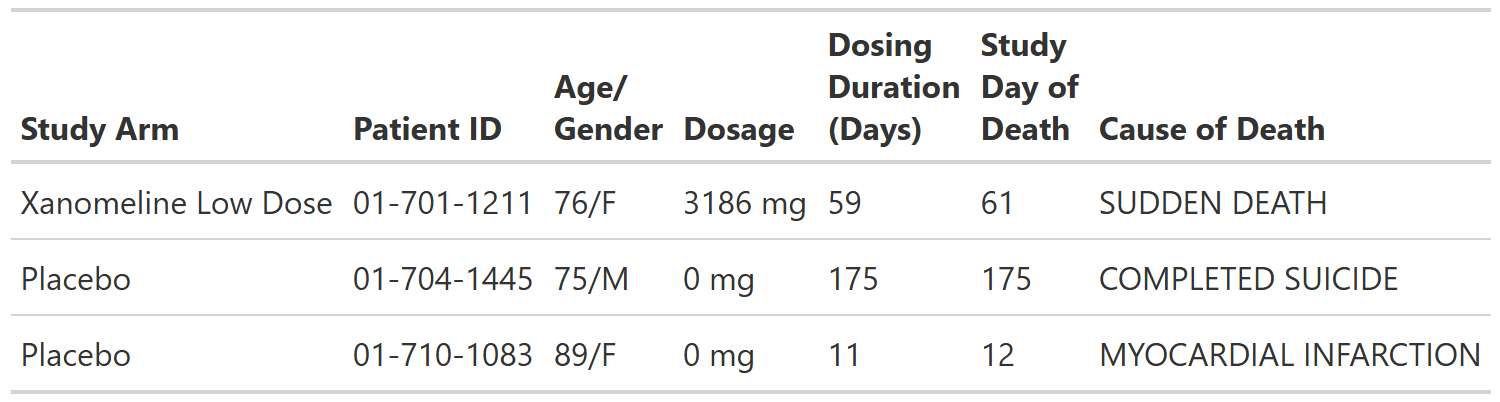
Deaths, Safety Population, Pooled Analysis (or Trial X)
FDA Table 08
table
FDA
safety
deaths
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adsl <- pharmaverseadam::adsl
adae <- pharmaverseadam::adae
# Pre-processing --------------------------------------------
adsl <- adsl |>
filter(SAFFL == "Y") # safety population
data <- adae |>
filter(
# safety population
SAFFL == "Y",
# deaths
DTHFL == "Y"
)Code
tbl_dthcaus <- data |>
tbl_hierarchical(
variables = DTHCAUS,
id = USUBJID,
by = TRT01A,
denominator = adsl,
overall_row = TRUE,
label = list("..ard_hierarchical_overall.." = "Death details", DTHCAUS = "**Deaths**")
) |>
sort_hierarchical() |>
modify_indent(columns = label, rows = variable == "DTHCAUS", indent = 4L)
tbl_aesdth <- data |>
filter(AESDTH == "Y") |>
tbl_hierarchical(
variables = AEDECOD,
id = USUBJID,
by = TRT01A,
denominator = adsl,
overall_row = TRUE,
label = list("..ard_hierarchical_overall.." = "Adverse events with an outcome of death", AEDECOD = "**Deaths**")
) |>
sort_hierarchical() |>
modify_indent(columns = label, rows = variable == "AEDECOD", indent = 4L)
tbl <- tbl_stack(list(tbl_dthcaus, tbl_aesdth))
tbl
[[1]]
[[1]]$tbl_hierarchical{cards} data frame: 33 x 13 group1 group1_level variable variable_level stat_name stat_label stat stat_fmt
1 <NA> TRT01A Placebo n n 86 86
2 <NA> TRT01A Placebo N N 254 254
3 <NA> TRT01A Placebo p % 0.339 33.9
4 <NA> TRT01A Xanomeli… n n 72 72
5 <NA> TRT01A Xanomeli… N N 254 254
6 <NA> TRT01A Xanomeli… p % 0.283 28.3
7 <NA> TRT01A Xanomeli… n n 96 96
8 <NA> TRT01A Xanomeli… N N 254 254
9 <NA> TRT01A Xanomeli… p % 0.378 37.8
10 TRT01A Placebo ..ard_hierarchical_overall.. TRUE n n 2 2ℹ 23 more rowsℹ Use `print(n = ...)` to see more rowsℹ 5 more variables: context, fmt_fun, warning, error, gts_column
[[2]]
[[2]]$tbl_hierarchical{cards} data frame: 33 x 13 group1 group1_level variable variable_level stat_name stat_label stat stat_fmt
1 <NA> TRT01A Placebo n n 86 86
2 <NA> TRT01A Placebo N N 254 254
3 <NA> TRT01A Placebo p % 0.339 33.9
4 <NA> TRT01A Xanomeli… n n 72 72
5 <NA> TRT01A Xanomeli… N N 254 254
6 <NA> TRT01A Xanomeli… p % 0.283 28.3
7 <NA> TRT01A Xanomeli… n n 96 96
8 <NA> TRT01A Xanomeli… N N 254 254
9 <NA> TRT01A Xanomeli… p % 0.378 37.8
10 TRT01A Placebo ..ard_hierarchical_overall.. TRUE n n 2 2ℹ 23 more rowsℹ Use `print(n = ...)` to see more rowsℹ 5 more variables: context, fmt_fun, warning, error, gts_column