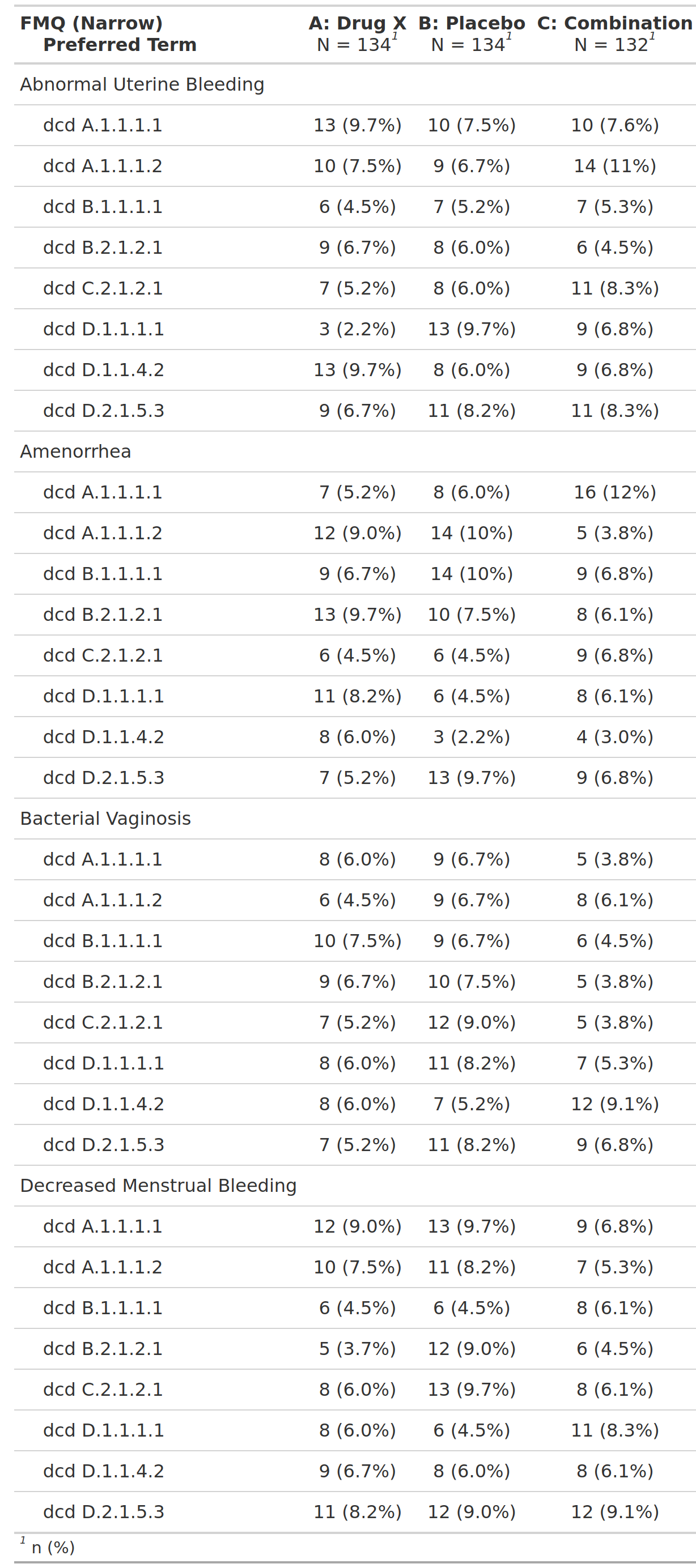
Subjects With Adverse Events by Organ System and OCMQ, Safety Population, Pooled Analysis (or Trial X)
FDA Table 17
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adae <- pharmaverseadam::adae |>
rename(OCMQ01SC = AEHLTCD, OCMQ01NAM = AEHLT)
adsl <- pharmaverseadam::adsl
set.seed(23)
adae$OCMQ01SC <- sample(c("BROAD", "NARROW"), size = nrow(adae), replace = TRUE)
# Pre-processing --------------------------------------------
adae <- adae |>
# safety population
filter(SAFFL == "Y") |>
# filter 0CMQ to truncate table
filter(OCMQ01NAM %in% c("HLT_0649", "HLT_0644", " HLT_0570", " HLT_0256", "HLT_0742", "HLT_0244", "HLT_0097", "HLT_0738"))
adsl <- adsl |>
# safety population
filter(SAFFL == "Y")Code
tbl <- adae |>
select(OCMQ01SC, TRT01A, OCMQ01NAM, AEBODSYS, USUBJID) |>
# setting an explicit level for NA values so empty strata combinations are shown.
mutate(across(everything(), ~ {
if (anyNA(.)) {
forcats::fct_na_value_to_level(as.factor(.), level = "<Missing>")
} else {
.
}
})) |>
tbl_strata(
strata = OCMQ01SC,
~ tbl_hierarchical(
.x,
by = TRT01A,
variables = c(AEBODSYS, OCMQ01NAM),
id = USUBJID,
denominator = adsl,
# variables to calculate rates for
include = c(OCMQ01NAM),
label = list(OCMQ01NAM ~ "OCMQ", AEBODSYS ~ "System Organ Class")
)
) |>
modify_missing_symbol("NA", columns = everything(), rows = row_type == "level")
tbl
$`OCMQ01SC="BROAD"`
$`OCMQ01SC="BROAD"`$tbl_hierarchical{cards} data frame: 63 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo AEBODSYS CARDIAC … OCMQ01NAM HLT_0644 hierarch… n n 0 0 <fn> stat_1ℹ 53 more rowsℹ Use `print(n = ...)` to see more rows
$`OCMQ01SC="NARROW"`
$`OCMQ01SC="NARROW"`$tbl_hierarchical{cards} data frame: 63 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> TRT01A Placebo tabulate n n 86 86 0 stat_1
2 <NA> <NA> TRT01A Placebo tabulate N N 254 254 0 stat_1
3 <NA> <NA> TRT01A Placebo tabulate p % 0.339 33.9 <fn> stat_1
4 <NA> <NA> TRT01A Xanomeli… tabulate n n 72 72 0 stat_2
5 <NA> <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_2
6 <NA> <NA> TRT01A Xanomeli… tabulate p % 0.283 28.3 <fn> stat_2
7 <NA> <NA> TRT01A Xanomeli… tabulate n n 96 96 0 stat_3
8 <NA> <NA> TRT01A Xanomeli… tabulate N N 254 254 0 stat_3
9 <NA> <NA> TRT01A Xanomeli… tabulate p % 0.378 37.8 <fn> stat_3
10 TRT01A Placebo AEBODSYS CARDIAC … OCMQ01NAM HLT_0644 hierarch… n n 2 2 <fn> stat_1ℹ 53 more rows
ℹ Use `print(n = ...)` to see more rows