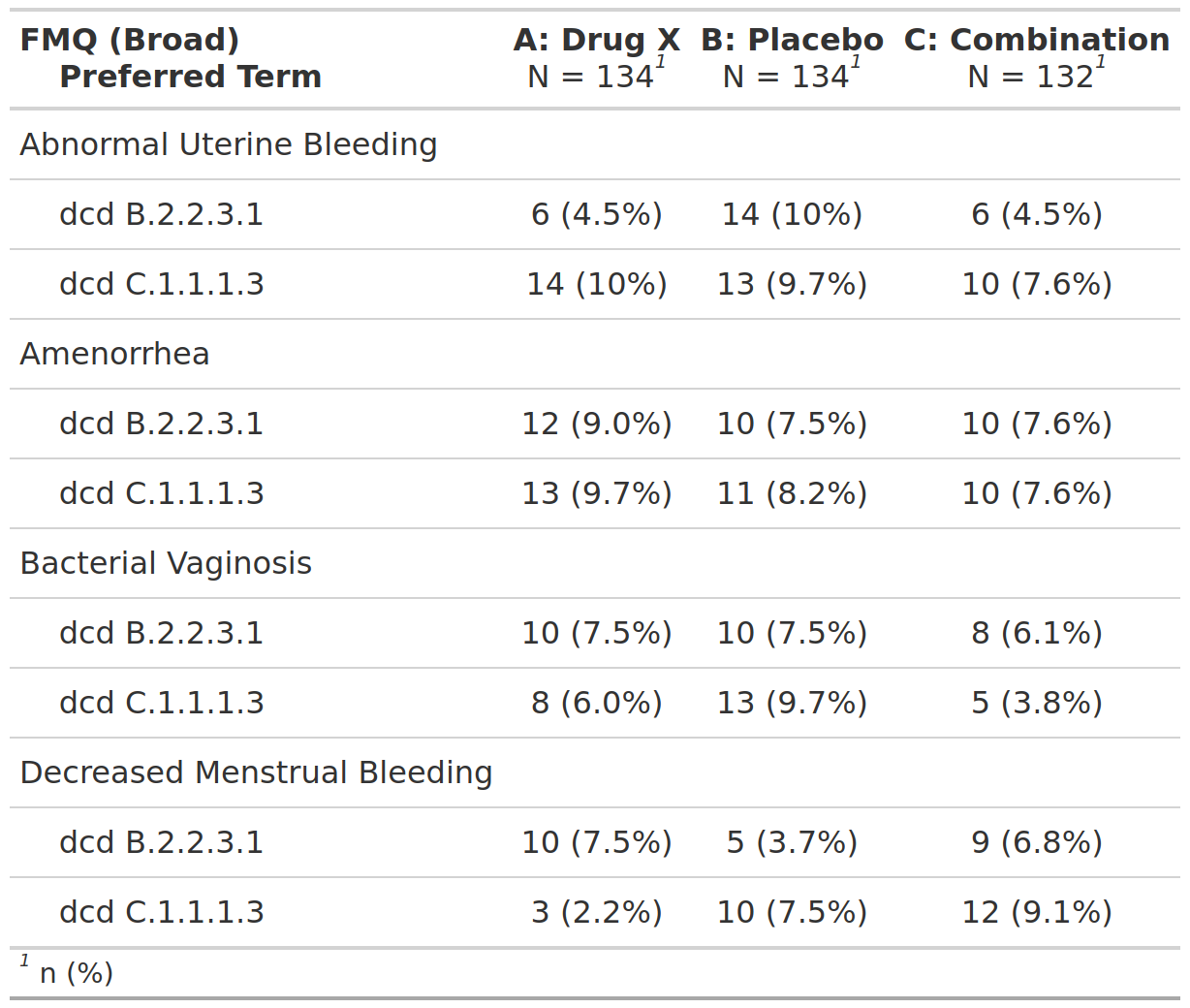
Summary Assessment of [Insert AE of Interest], Safety Population, Pooled Analysis (or Trial X)
FDA Table 18
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(cardx)
library(gtsummary)
adsl <- pharmaverseadam::adsl
adae <- pharmaverseadam::adae
set.seed(1)
adae$AESIFL <- ifelse(adae$AESOC %in% c("VASCULAR DISORDERS"), "Y", "N")
# Pre-processing --------------------------------------------
adsl <- adsl |>
# safety population
filter(SAFFL == "Y")
data <- adae |>
filter(
# safety population
SAFFL == "Y",
# AESI assessment
AESIFL == "Y"
)
# dataset of AE flag variable counts by subject
data_ae_fl <-
adsl |>
select(USUBJID, TRT01A) |>
# create subject-level flags from adae data
left_join(
data |>
select(USUBJID, AESER, AESDTH, AESLIFE, AESHOSP, AESDISAB, AESCONG, AEACN) |>
mutate(
# Serious AE
ae_ser = any(AESER == "Y"),
# Serious AE leading to death
ae_ser_death = any(AESER == "Y" & AESDTH == "Y"),
ae_ser_life = any(AESER == "Y" & AESLIFE == "Y"),
ae_ser_hosp = any(AESER == "Y" & AESHOSP == "Y"),
ae_ser_disab = any(AESER == "Y" & AESDISAB == "Y"),
ae_ser_cong = any(AESER == "Y" & AESCONG == "Y"),
# AE resulting in discontinuation
ae_withdraw = any(AEACN == "DRUG WITHDRAWN"),
.by = USUBJID
),
by = "USUBJID"
) |>
distinct(USUBJID, .keep_all = TRUE) |>
# add number of AEs
left_join(
data |> summarise(.by = USUBJID, ae_count = n()),
by = "USUBJID",
relationship = "one-to-one"
)Code
## AE grouping flags section --------
tbl_ae_gp <- data |>
tbl_hierarchical(
variables = AEDECOD,
id = USUBJID,
denominator = adsl,
by = TRT01A,
overall_row = TRUE,
label = list(
..ard_hierarchical_overall.. = "[Insert AE of Interest]",
AEDECOD = "AE of Interest Assessment"
)
) |>
modify_indent(columns = label, rows = variable != "..ard_hierarchical_overall..", indent = 4L)
## Maximum severity section ---------
tbl_ae_sev <- data |>
ard_tabulate_max(variables = AESEV, id = USUBJID, by = TRT01A, denominator = adsl, quiet = TRUE) |>
tbl_ard_summary(by = TRT01A, label = list(AESEV = "Maximum severity"))
## Events section -------------------
tbl_ae_fl <- data_ae_fl |>
tbl_summary(
by = "TRT01A",
include = c(ae_ser, ae_ser_death, ae_ser_life, ae_ser_hosp, ae_ser_disab, ae_ser_cong, ae_withdraw),
missing = "no",
percent = adsl,
label = list(
ae_ser = "SAE",
ae_ser_death = "Death",
ae_ser_life = "Life-threatening",
ae_ser_hosp = "Initial or prolonged hospitalization",
ae_ser_disab = "Disability or permanent damage",
ae_ser_cong = "Congenital anomaly or birth defect",
ae_withdraw = "Resulting in treatment discontinuation"
)
) |>
modify_indent(columns = label, rows = !variable %in% c("ae_ser", "ae_withdraw"), indent = 4L)
## Build table ----------------------
tbl <- tbl_stack(list(tbl_ae_gp, tbl_ae_sev, tbl_ae_fl), quiet = TRUE)
tbl
[[1]]
[[1]]$tbl_hierarchical{cards} data frame: 63 x 13 group1 group1_level variable variable_level stat_name stat_label stat stat_fmt
1 <NA> TRT01A Placebo n n 86 86
2 <NA> TRT01A Placebo N N 254 254
3 <NA> TRT01A Placebo p % 0.339 33.9
4 <NA> TRT01A Xanomeli… n n 72 72
5 <NA> TRT01A Xanomeli… N N 254 254
6 <NA> TRT01A Xanomeli… p % 0.283 28.3
7 <NA> TRT01A Xanomeli… n n 96 96
8 <NA> TRT01A Xanomeli… N N 254 254
9 <NA> TRT01A Xanomeli… p % 0.378 37.8
10 TRT01A Placebo ..ard_hierarchical_overall.. TRUE n n 3 3ℹ 53 more rowsℹ Use `print(n = ...)` to see more rowsℹ 5 more variables: context, fmt_fun, warning, error, gts_column
[[2]]
[[2]]$tbl_ard_summary{cards} data frame: 20 x 12 group1 group1_level variable variable_level stat_name stat_label stat
1 TRT01A Placebo AESEV MILD n n 1
2 TRT01A Placebo AESEV MILD N N 86
3 TRT01A Placebo AESEV MILD p % 0.012
4 TRT01A Placebo AESEV MODERATE n n 2
5 TRT01A Placebo AESEV MODERATE N N 86
6 TRT01A Placebo AESEV MODERATE p % 0.023
7 TRT01A Xanomeli… AESEV MILD n n 0
8 TRT01A Xanomeli… AESEV MILD N N 72
9 TRT01A Xanomeli… AESEV MILD p % 0
10 TRT01A Xanomeli… AESEV MODERATE n n 1
11 TRT01A Xanomeli… AESEV MODERATE N N 72
12 TRT01A Xanomeli… AESEV MODERATE p % 0.014
13 TRT01A Xanomeli… AESEV MILD n n 3
14 TRT01A Xanomeli… AESEV MILD N N 96
15 TRT01A Xanomeli… AESEV MILD p % 0.031
16 TRT01A Xanomeli… AESEV MODERATE n n 1
17 TRT01A Xanomeli… AESEV MODERATE N N 96
18 TRT01A Xanomeli… AESEV MODERATE p % 0.01
19 <NA> AESEV label Variable… Maximum …
20 <NA> AESEV class Variable… logicalℹ 5 more variables: context, fmt_fun, warning, error, gts_column
[[3]]
[[3]]$tbl_summary{cards} data frame: 194 x 12 group1 group1_level variable variable_level stat_name stat_label stat
1 TRT01A Placebo ae_ser TRUE n n 0
2 TRT01A Placebo ae_ser TRUE N N 86
3 TRT01A Placebo ae_ser TRUE p % 0
4 TRT01A Placebo ae_ser_death TRUE n n 0
5 TRT01A Placebo ae_ser_death TRUE N N 86
6 TRT01A Placebo ae_ser_death TRUE p % 0
7 TRT01A Placebo ae_ser_life TRUE n n 0
8 TRT01A Placebo ae_ser_life TRUE N N 86
9 TRT01A Placebo ae_ser_life TRUE p % 0
10 TRT01A Placebo ae_ser_hosp TRUE n n 0ℹ 184 more rowsℹ Use `print(n = ...)` to see more rowsℹ 5 more variables: context, fmt_fun, warning, error, gts_column