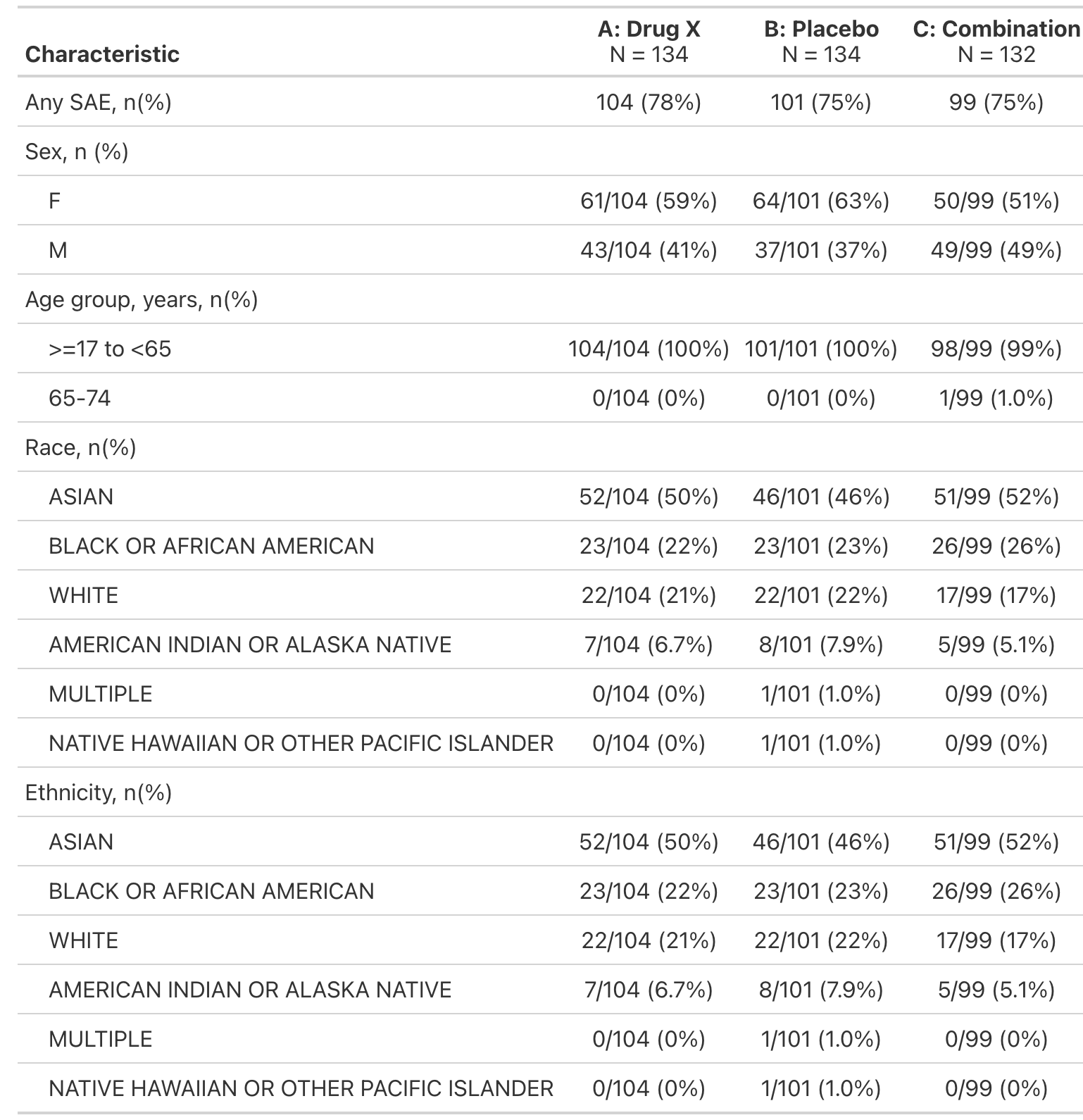
FDA Table 21
Overview of Serious Adverse Events by Demographic Subgroup, Safety Population, Pooled Analysis (or Trial X)
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adae <- random.cdisc.data::cadae
adsl <- random.cdisc.data::cadsl
adsl <- adsl |>
filter(SAFFL == "Y")
data <- adae |>
filter(
SAFFL == "Y",
TRTEMFL == "Y",
AESER == "Y"
) |>
mutate(
SEXGR = "Sex, n (%)",
SEXGR1 = as.factor(SEX),
AGEGR = "Age group, years, n(%)",
AGEGR1 = factor(
dplyr::case_when(
AGE >= 17 & AGE < 65 ~ ">=17 to <65",
AGE >= 65 & AGE < 75 ~ "65-74",
AGE >= 75 ~ ">=75"
),
levels = c(">=17 to <65", "65-74", ">=75")
),
RACEGR = "Race, n(%)",
RACEGR1 = as.factor(RACE),
ETHNICGR = "Ethnicity, n(%)",
ETHNICGR1 = as.factor(RACE)
)
data_any_sae <- adae |>
filter(AESER == "Y") |>
mutate(AESER = "Any SAE, n(%)")Code
tbl_any_sae <- tbl_hierarchical(
data = data_any_sae,
denominator = adsl,
id = "USUBJID",
by = "ARM",
variables = "AESER",
statistic = ~"{n} ({p}%)",
label = AESER ~ "Characteristic"
)
tbl_sex <- tbl_hierarchical(
data = data,
denominator = data_any_sae |> slice_head(by = c(USUBJID)),
id = "USUBJID",
by = "ARM",
variables = c(SEXGR, SEXGR1),
include = SEXGR1,
statistic = ~"{n}/{N} ({p}%)"
)
tbl_agegr1 <- tbl_hierarchical(
data = data,
denominator = data_any_sae |> slice_head(by = c(USUBJID)),
id = "USUBJID",
by = "ARM",
variables = c(AGEGR, AGEGR1),
include = AGEGR1,
statistic = ~"{n}/{N} ({p}%)"
)
tbl_race <- tbl_hierarchical(
data = data,
denominator = data_any_sae |> slice_head(by = c(USUBJID)),
id = "USUBJID",
by = "ARM",
variables = c(RACEGR, RACEGR1),
include = RACEGR1,
statistic = ~"{n}/{N} ({p}%)"
)
tbl_ethnic <- tbl_hierarchical(
data = data,
denominator = data_any_sae |> slice_head(by = c(USUBJID)),
id = "USUBJID",
by = "ARM",
variables = c(ETHNICGR, ETHNICGR1),
include = ETHNICGR1,
statistic = ~"{n}/{N} ({p}%)"
)
tbl <- list(tbl_any_sae, tbl_sex, tbl_agegr1, tbl_race, tbl_ethnic) |>
tbl_stack() |>
modify_column_indent("label", rows = !(variable %in% c("..ard_hierarchical_overall..", "AESER", "SEXGR", "AGEGR", "RACEGR", "ETHNICGR"))) |>
remove_footnote_header(columns = everything())
tbl| Characteristic |
A: Drug X N = 134 |
B: Placebo N = 134 |
C: Combination N = 132 |
|---|---|---|---|
| Any SAE, n(%) | 104 (78%) | 101 (75%) | 99 (75%) |
| Sex, n (%) | |||
| F | 61/104 (59%) | 64/101 (63%) | 50/99 (51%) |
| M | 43/104 (41%) | 37/101 (37%) | 49/99 (49%) |
| Age group, years, n(%) | |||
| >=17 to <65 | 104/104 (100%) | 101/101 (100%) | 98/99 (99%) |
| 65-74 | 0/104 (0%) | 0/101 (0%) | 1/99 (1.0%) |
| Race, n(%) | |||
| ASIAN | 52/104 (50%) | 46/101 (46%) | 51/99 (52%) |
| BLACK OR AFRICAN AMERICAN | 23/104 (22%) | 23/101 (23%) | 26/99 (26%) |
| WHITE | 22/104 (21%) | 22/101 (22%) | 17/99 (17%) |
| AMERICAN INDIAN OR ALASKA NATIVE | 7/104 (6.7%) | 8/101 (7.9%) | 5/99 (5.1%) |
| MULTIPLE | 0/104 (0%) | 1/101 (1.0%) | 0/99 (0%) |
| NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | 0/104 (0%) | 1/101 (1.0%) | 0/99 (0%) |
| Ethnicity, n(%) | |||
| ASIAN | 52/104 (50%) | 46/101 (46%) | 51/99 (52%) |
| BLACK OR AFRICAN AMERICAN | 23/104 (22%) | 23/101 (23%) | 26/99 (26%) |
| WHITE | 22/104 (21%) | 22/101 (22%) | 17/99 (17%) |
| AMERICAN INDIAN OR ALASKA NATIVE | 7/104 (6.7%) | 8/101 (7.9%) | 5/99 (5.1%) |
| MULTIPLE | 0/104 (0%) | 1/101 (1.0%) | 0/99 (0%) |
| NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | 0/104 (0%) | 1/101 (1.0%) | 0/99 (0%) |
[[1]]
[[1]]$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESER Any SAE,… hierarch… n n 104 104 <fn> stat_1
11 ARM A: Drug X AESER Any SAE,… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESER Any SAE,… hierarch… p % 0.776 78 <fn> stat_1
13 ARM B: Place… AESER Any SAE,… hierarch… n n 101 101 <fn> stat_2
14 ARM B: Place… AESER Any SAE,… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESER Any SAE,… hierarch… p % 0.754 75 <fn> stat_2
16 ARM C: Combi… AESER Any SAE,… hierarch… n n 99 99 <fn> stat_3
17 ARM C: Combi… AESER Any SAE,… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESER Any SAE,… hierarch… p % 0.75 75 <fn> stat_3
[[2]]
[[2]]$tbl_hierarchical{cards} data frame: 27 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> ARM A: Drug X categori… n n 104 104 0 stat_1
2 <NA> <NA> ARM A: Drug X categori… N N 304 304 0 stat_1
3 <NA> <NA> ARM A: Drug X categori… p % 0.342 34.2 <fn> stat_1
4 <NA> <NA> ARM B: Place… categori… n n 101 101 0 stat_2
5 <NA> <NA> ARM B: Place… categori… N N 304 304 0 stat_2
6 <NA> <NA> ARM B: Place… categori… p % 0.332 33.2 <fn> stat_2
7 <NA> <NA> ARM C: Combi… categori… n n 99 99 0 stat_3
8 <NA> <NA> ARM C: Combi… categori… N N 304 304 0 stat_3
9 <NA> <NA> ARM C: Combi… categori… p % 0.326 32.6 <fn> stat_3
10 ARM A: Drug X SEXGR Sex, n (… SEXGR1 F hierarch… n n 61 61 <fn> stat_1
11 ARM A: Drug X SEXGR Sex, n (… SEXGR1 F hierarch… N N 104 104 <fn> stat_1
12 ARM A: Drug X SEXGR Sex, n (… SEXGR1 F hierarch… p % 0.587 59 <fn> stat_1
13 ARM B: Place… SEXGR Sex, n (… SEXGR1 F hierarch… n n 64 64 <fn> stat_2
14 ARM B: Place… SEXGR Sex, n (… SEXGR1 F hierarch… N N 101 101 <fn> stat_2
15 ARM B: Place… SEXGR Sex, n (… SEXGR1 F hierarch… p % 0.634 63 <fn> stat_2
16 ARM C: Combi… SEXGR Sex, n (… SEXGR1 F hierarch… n n 50 50 <fn> stat_3
17 ARM C: Combi… SEXGR Sex, n (… SEXGR1 F hierarch… N N 99 99 <fn> stat_3
18 ARM C: Combi… SEXGR Sex, n (… SEXGR1 F hierarch… p % 0.505 51 <fn> stat_3
19 ARM A: Drug X SEXGR Sex, n (… SEXGR1 M hierarch… n n 43 43 <fn> stat_1
20 ARM A: Drug X SEXGR Sex, n (… SEXGR1 M hierarch… N N 104 104 <fn> stat_1
21 ARM A: Drug X SEXGR Sex, n (… SEXGR1 M hierarch… p % 0.413 41 <fn> stat_1
22 ARM B: Place… SEXGR Sex, n (… SEXGR1 M hierarch… n n 37 37 <fn> stat_2
23 ARM B: Place… SEXGR Sex, n (… SEXGR1 M hierarch… N N 101 101 <fn> stat_2
24 ARM B: Place… SEXGR Sex, n (… SEXGR1 M hierarch… p % 0.366 37 <fn> stat_2
25 ARM C: Combi… SEXGR Sex, n (… SEXGR1 M hierarch… n n 49 49 <fn> stat_3
26 ARM C: Combi… SEXGR Sex, n (… SEXGR1 M hierarch… N N 99 99 <fn> stat_3
27 ARM C: Combi… SEXGR Sex, n (… SEXGR1 M hierarch… p % 0.495 49 <fn> stat_3
[[3]]
[[3]]$tbl_hierarchical{cards} data frame: 27 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> ARM A: Drug X categori… n n 104 104 0 stat_1
2 <NA> <NA> ARM A: Drug X categori… N N 304 304 0 stat_1
3 <NA> <NA> ARM A: Drug X categori… p % 0.342 34.2 <fn> stat_1
4 <NA> <NA> ARM B: Place… categori… n n 101 101 0 stat_2
5 <NA> <NA> ARM B: Place… categori… N N 304 304 0 stat_2
6 <NA> <NA> ARM B: Place… categori… p % 0.332 33.2 <fn> stat_2
7 <NA> <NA> ARM C: Combi… categori… n n 99 99 0 stat_3
8 <NA> <NA> ARM C: Combi… categori… N N 304 304 0 stat_3
9 <NA> <NA> ARM C: Combi… categori… p % 0.326 32.6 <fn> stat_3
10 ARM A: Drug X AGEGR Age grou… AGEGR1 >=17 to … hierarch… n n 104 104 <fn> stat_1
11 ARM A: Drug X AGEGR Age grou… AGEGR1 >=17 to … hierarch… N N 104 104 <fn> stat_1
12 ARM A: Drug X AGEGR Age grou… AGEGR1 >=17 to … hierarch… p % 1 100 <fn> stat_1
13 ARM B: Place… AGEGR Age grou… AGEGR1 >=17 to … hierarch… n n 101 101 <fn> stat_2
14 ARM B: Place… AGEGR Age grou… AGEGR1 >=17 to … hierarch… N N 101 101 <fn> stat_2
15 ARM B: Place… AGEGR Age grou… AGEGR1 >=17 to … hierarch… p % 1 100 <fn> stat_2
16 ARM C: Combi… AGEGR Age grou… AGEGR1 >=17 to … hierarch… n n 98 98 <fn> stat_3
17 ARM C: Combi… AGEGR Age grou… AGEGR1 >=17 to … hierarch… N N 99 99 <fn> stat_3
18 ARM C: Combi… AGEGR Age grou… AGEGR1 >=17 to … hierarch… p % 0.99 99 <fn> stat_3
19 ARM A: Drug X AGEGR Age grou… AGEGR1 65-74 hierarch… n n 0 0 <fn> stat_1
20 ARM A: Drug X AGEGR Age grou… AGEGR1 65-74 hierarch… N N 104 104 <fn> stat_1
21 ARM A: Drug X AGEGR Age grou… AGEGR1 65-74 hierarch… p % 0 0 <fn> stat_1
22 ARM B: Place… AGEGR Age grou… AGEGR1 65-74 hierarch… n n 0 0 <fn> stat_2
23 ARM B: Place… AGEGR Age grou… AGEGR1 65-74 hierarch… N N 101 101 <fn> stat_2
24 ARM B: Place… AGEGR Age grou… AGEGR1 65-74 hierarch… p % 0 0 <fn> stat_2
25 ARM C: Combi… AGEGR Age grou… AGEGR1 65-74 hierarch… n n 1 1 <fn> stat_3
26 ARM C: Combi… AGEGR Age grou… AGEGR1 65-74 hierarch… N N 99 99 <fn> stat_3
27 ARM C: Combi… AGEGR Age grou… AGEGR1 65-74 hierarch… p % 0.01 1.0 <fn> stat_3
[[4]]
[[4]]$tbl_hierarchical{cards} data frame: 63 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> ARM A: Drug X categori… n n 104 104 0 stat_1
2 <NA> <NA> ARM A: Drug X categori… N N 304 304 0 stat_1
3 <NA> <NA> ARM A: Drug X categori… p % 0.342 34.2 <fn> stat_1
4 <NA> <NA> ARM B: Place… categori… n n 101 101 0 stat_2
5 <NA> <NA> ARM B: Place… categori… N N 304 304 0 stat_2
6 <NA> <NA> ARM B: Place… categori… p % 0.332 33.2 <fn> stat_2
7 <NA> <NA> ARM C: Combi… categori… n n 99 99 0 stat_3
8 <NA> <NA> ARM C: Combi… categori… N N 304 304 0 stat_3
9 <NA> <NA> ARM C: Combi… categori… p % 0.326 32.6 <fn> stat_3
10 ARM A: Drug X RACEGR Race, n(… RACEGR1 ASIAN hierarch… n n 52 52 <fn> stat_1
11 ARM A: Drug X RACEGR Race, n(… RACEGR1 ASIAN hierarch… N N 104 104 <fn> stat_1
12 ARM A: Drug X RACEGR Race, n(… RACEGR1 ASIAN hierarch… p % 0.5 50 <fn> stat_1
13 ARM B: Place… RACEGR Race, n(… RACEGR1 ASIAN hierarch… n n 46 46 <fn> stat_2
14 ARM B: Place… RACEGR Race, n(… RACEGR1 ASIAN hierarch… N N 101 101 <fn> stat_2
15 ARM B: Place… RACEGR Race, n(… RACEGR1 ASIAN hierarch… p % 0.455 46 <fn> stat_2
16 ARM C: Combi… RACEGR Race, n(… RACEGR1 ASIAN hierarch… n n 51 51 <fn> stat_3
17 ARM C: Combi… RACEGR Race, n(… RACEGR1 ASIAN hierarch… N N 99 99 <fn> stat_3
18 ARM C: Combi… RACEGR Race, n(… RACEGR1 ASIAN hierarch… p % 0.515 52 <fn> stat_3
19 ARM A: Drug X RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… n n 23 23 <fn> stat_1
20 ARM A: Drug X RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… N N 104 104 <fn> stat_1
21 ARM A: Drug X RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… p % 0.221 22 <fn> stat_1
22 ARM B: Place… RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… n n 23 23 <fn> stat_2
23 ARM B: Place… RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… N N 101 101 <fn> stat_2
24 ARM B: Place… RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… p % 0.228 23 <fn> stat_2
25 ARM C: Combi… RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… n n 26 26 <fn> stat_3
26 ARM C: Combi… RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… N N 99 99 <fn> stat_3
27 ARM C: Combi… RACEGR Race, n(… RACEGR1 BLACK OR… hierarch… p % 0.263 26 <fn> stat_3
28 ARM A: Drug X RACEGR Race, n(… RACEGR1 WHITE hierarch… n n 22 22 <fn> stat_1
29 ARM A: Drug X RACEGR Race, n(… RACEGR1 WHITE hierarch… N N 104 104 <fn> stat_1
30 ARM A: Drug X RACEGR Race, n(… RACEGR1 WHITE hierarch… p % 0.212 21 <fn> stat_1
31 ARM B: Place… RACEGR Race, n(… RACEGR1 WHITE hierarch… n n 22 22 <fn> stat_2
32 ARM B: Place… RACEGR Race, n(… RACEGR1 WHITE hierarch… N N 101 101 <fn> stat_2
33 ARM B: Place… RACEGR Race, n(… RACEGR1 WHITE hierarch… p % 0.218 22 <fn> stat_2
34 ARM C: Combi… RACEGR Race, n(… RACEGR1 WHITE hierarch… n n 17 17 <fn> stat_3
35 ARM C: Combi… RACEGR Race, n(… RACEGR1 WHITE hierarch… N N 99 99 <fn> stat_3
36 ARM C: Combi… RACEGR Race, n(… RACEGR1 WHITE hierarch… p % 0.172 17 <fn> stat_3
37 ARM A: Drug X RACEGR Race, n(… RACEGR1 AMERICAN… hierarch… n n 7 7 <fn> stat_1
38 ARM A: Drug X RACEGR Race, n(… RACEGR1 AMERICAN… hierarch… N N 104 104 <fn> stat_1
39 ARM A: Drug X RACEGR Race, n(… RACEGR1 AMERICAN… hierarch… p % 0.067 6.7 <fn> stat_1
40 ARM B: Place… RACEGR Race, n(… RACEGR1 AMERICAN… hierarch… n n 8 8 <fn> stat_2ℹ 23 more rowsℹ Use `print(n = ...)` to see more rows
[[5]]
[[5]]$tbl_hierarchical{cards} data frame: 63 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> ARM A: Drug X categori… n n 104 104 0 stat_1
2 <NA> <NA> ARM A: Drug X categori… N N 304 304 0 stat_1
3 <NA> <NA> ARM A: Drug X categori… p % 0.342 34.2 <fn> stat_1
4 <NA> <NA> ARM B: Place… categori… n n 101 101 0 stat_2
5 <NA> <NA> ARM B: Place… categori… N N 304 304 0 stat_2
6 <NA> <NA> ARM B: Place… categori… p % 0.332 33.2 <fn> stat_2
7 <NA> <NA> ARM C: Combi… categori… n n 99 99 0 stat_3
8 <NA> <NA> ARM C: Combi… categori… N N 304 304 0 stat_3
9 <NA> <NA> ARM C: Combi… categori… p % 0.326 32.6 <fn> stat_3
10 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… n n 52 52 <fn> stat_1
11 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… N N 104 104 <fn> stat_1
12 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… p % 0.5 50 <fn> stat_1
13 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… n n 46 46 <fn> stat_2
14 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… N N 101 101 <fn> stat_2
15 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… p % 0.455 46 <fn> stat_2
16 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… n n 51 51 <fn> stat_3
17 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… N N 99 99 <fn> stat_3
18 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 ASIAN hierarch… p % 0.515 52 <fn> stat_3
19 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… n n 23 23 <fn> stat_1
20 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… N N 104 104 <fn> stat_1
21 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… p % 0.221 22 <fn> stat_1
22 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… n n 23 23 <fn> stat_2
23 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… N N 101 101 <fn> stat_2
24 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… p % 0.228 23 <fn> stat_2
25 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… n n 26 26 <fn> stat_3
26 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… N N 99 99 <fn> stat_3
27 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 BLACK OR… hierarch… p % 0.263 26 <fn> stat_3
28 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… n n 22 22 <fn> stat_1
29 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… N N 104 104 <fn> stat_1
30 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… p % 0.212 21 <fn> stat_1
31 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… n n 22 22 <fn> stat_2
32 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… N N 101 101 <fn> stat_2
33 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… p % 0.218 22 <fn> stat_2
34 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… n n 17 17 <fn> stat_3
35 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… N N 99 99 <fn> stat_3
36 ARM C: Combi… ETHNICGR Ethnicit… ETHNICGR1 WHITE hierarch… p % 0.172 17 <fn> stat_3
37 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 AMERICAN… hierarch… n n 7 7 <fn> stat_1
38 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 AMERICAN… hierarch… N N 104 104 <fn> stat_1
39 ARM A: Drug X ETHNICGR Ethnicit… ETHNICGR1 AMERICAN… hierarch… p % 0.067 6.7 <fn> stat_1
40 ARM B: Place… ETHNICGR Ethnicit… ETHNICGR1 AMERICAN… hierarch… n n 8 8 <fn> stat_2ℹ 23 more rows
ℹ Use `print(n = ...)` to see more rows