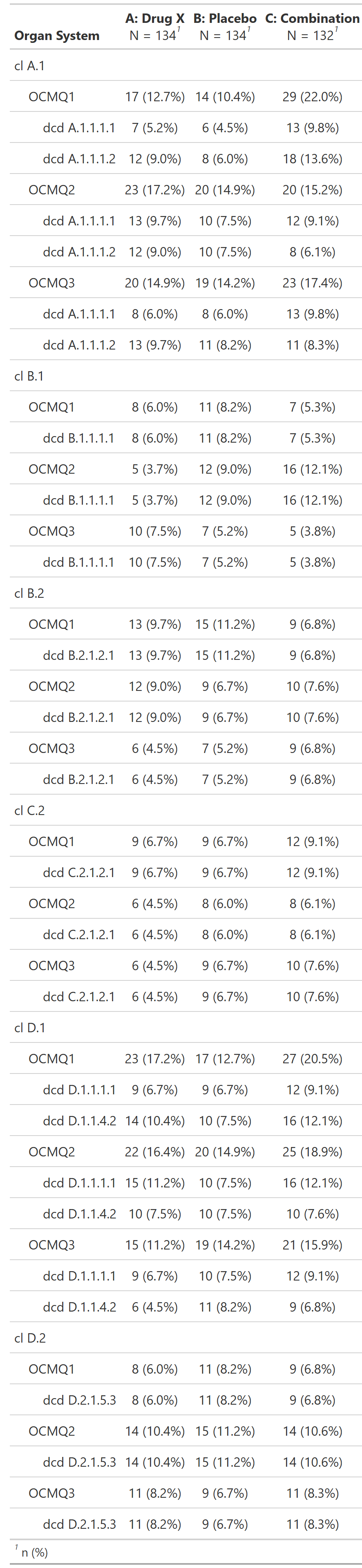
Subjects With Serious Adverse Events by Organ System, OCMQ (Narrow) and Preferred Term, Safety Population, Pooled Analysis (or Trial X)
FDA Table 29
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adae <- pharmaverseadam::adae
adsl <- pharmaverseadam::adsl
set.seed(1)
adae <- adae |>
rename(OCMQ01SC = AEHLTCD) |>
mutate(
AESER = sample(c("Y", "N"), size = nrow(adae), replace = TRUE),
OCMQ01NAM = sample(c("OCMQ1", "OCMQ2", "OCMQ3"), size = nrow(adae), replace = TRUE)
) |>
# truncating table for demonstration
filter(AESOC == "VASCULAR DISORDERS")
adae$OCMQ01SC[is.na(adae$OCMQ01SC)] <- "NARROW"
# Pre-processing --------------------------------------------
adsl <- adsl |>
# safety population
filter(
SAFFL == "Y"
)
data <- adae |>
filter(
# safety population
SAFFL == "Y",
# serious AEs
AESER == "Y",
# treatment-emergent
TRTEMFL == "Y",
# narrow OCMQ scope
OCMQ01SC == "NARROW"
){cards} data frame: 57 x 15 group1 group1_level group2 group2_level group3 group3_level variable variable_level context stat_name stat_label stat fmt_fun warning error
1 <NA> <NA> <NA> TRT01A Placebo tabulate n n 86 0
2 <NA> <NA> <NA> TRT01A Placebo tabulate N N 254 0
3 <NA> <NA> <NA> TRT01A Placebo tabulate p % 0.339 <fn>
4 <NA> <NA> <NA> TRT01A Xanomeli… tabulate n n 72 0
5 <NA> <NA> <NA> TRT01A Xanomeli… tabulate N N 254 0
6 <NA> <NA> <NA> TRT01A Xanomeli… tabulate p % 0.283 <fn>
7 <NA> <NA> <NA> TRT01A Xanomeli… tabulate n n 96 0
8 <NA> <NA> <NA> TRT01A Xanomeli… tabulate N N 254 0
9 <NA> <NA> <NA> TRT01A Xanomeli… tabulate p % 0.378 <fn>
10 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ1 hierarch… n n 1 0
11 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ1 hierarch… N N 86 0
12 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ1 hierarch… p % 0.012 <fn>
13 TRT01A Xanomeli… AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ1 hierarch… n n 1 0
14 TRT01A Xanomeli… AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ1 hierarch… N N 96 0
15 TRT01A Xanomeli… AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ1 hierarch… p % 0.01 <fn>
16 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ1 AEDECOD HYPOTENS… hierarch… n n 1 0
17 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ1 AEDECOD HYPOTENS… hierarch… N N 86 0
18 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ1 AEDECOD HYPOTENS… hierarch… p % 0.012 <fn>
19 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ1 AEDECOD HYPOTENS… hierarch… n n 1 0
20 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ1 AEDECOD HYPOTENS… hierarch… N N 96 0
21 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ1 AEDECOD HYPOTENS… hierarch… p % 0.01 <fn>
22 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ2 hierarch… n n 1 0
23 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ2 hierarch… N N 86 0
24 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ2 hierarch… p % 0.012 <fn>
25 TRT01A Xanomeli… AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ2 hierarch… n n 1 0
26 TRT01A Xanomeli… AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ2 hierarch… N N 96 0
27 TRT01A Xanomeli… AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ2 hierarch… p % 0.01 <fn>
28 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HOT FLUSH hierarch… n n 0 0
29 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HOT FLUSH hierarch… N N 86 0
30 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HOT FLUSH hierarch… p % 0 <fn>
31 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HOT FLUSH hierarch… n n 1 0
32 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HOT FLUSH hierarch… N N 96 0
33 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HOT FLUSH hierarch… p % 0.01 <fn>
34 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HYPOTENS… hierarch… n n 1 0
35 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HYPOTENS… hierarch… N N 86 0
36 TRT01A Placebo AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HYPOTENS… hierarch… p % 0.012 <fn>
37 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HYPOTENS… hierarch… n n 0 0
38 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HYPOTENS… hierarch… N N 96 0
39 TRT01A Xanomeli… AEBODSYS VASCULAR… OCMQ01NAM OCMQ2 AEDECOD HYPOTENS… hierarch… p % 0 <fn>
40 TRT01A Placebo AEBODSYS VASCULAR… <NA> OCMQ01NAM OCMQ3 hierarch… n n 2 0 ℹ 17 more rowsℹ Use `print(n = ...)` to see more rows