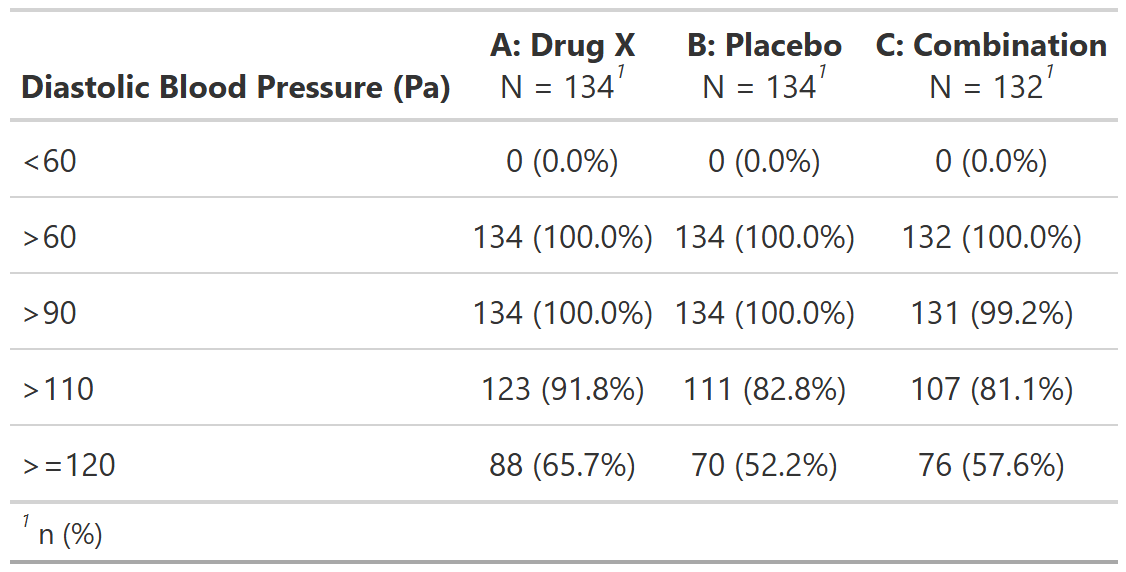
Subjects With Maximum Postbaseline Diastolic Blood Pressure by Category of Blood Pressure, Safety Population, Pooled Analysis (or Trial X)
FDA Table 37
table
FDA
safety
vital signs
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adsl <- pharmaverseadam::adsl
advs <- pharmaverseadam::advs
# Pre-processing --------------------------------------------
adsl <- adsl |>
filter(SAFFL == "Y") # safety population
data <- advs |>
filter(
# safety population
SAFFL == "Y",
# diastolic blood pressure
PARAMCD == "DIABP",
# post-baseline visits
AVISITN >= 2
) |>
# analyze maximum values of each subject
slice_max(AVAL, n = 1L, by = USUBJID) |>
# define analysis value cutoffs
mutate(
L60 = AVAL < 60, # DIABP <60
G60 = AVAL > 60, # DIABP >60
G90 = AVAL > 90, # DIABP >90
G110 = AVAL > 110, # DIABP >110
GE120 = AVAL >= 120 # DIABP >=120
)Code
ard <- bind_ard(
ard_tabulate_value(
data,
variables = c(L60, G60, G90, G110, GE120),
by = TRT01A,
statistic = ~ c("n", "p"),
# denominator values are number of subjects in arm with BP data
denominator = data |> distinct(USUBJID, TRT01A)
),
# ARD for header N values
ard_tabulate(adsl, variables = TRT01A)
)
ard{cards} data frame: 39 x 11 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error
1 TRT01A Placebo L60 TRUE tabulate… n n 1 0
2 TRT01A Placebo L60 TRUE tabulate… p % 0.012 <fn>
3 TRT01A Placebo G60 TRUE tabulate… n n 151 0
4 TRT01A Placebo G60 TRUE tabulate… p % 1.798 <fn>
5 TRT01A Placebo G90 TRUE tabulate… n n 36 0
6 TRT01A Placebo G90 TRUE tabulate… p % 0.429 <fn>
7 TRT01A Placebo G110 TRUE tabulate… n n 1 0
8 TRT01A Placebo G110 TRUE tabulate… p % 0.012 <fn>
9 TRT01A Placebo GE120 TRUE tabulate… n n 0 0
10 TRT01A Placebo GE120 TRUE tabulate… p % 0 <fn>
11 TRT01A Xanomeli… L60 TRUE tabulate… n n 0 0
12 TRT01A Xanomeli… L60 TRUE tabulate… p % 0 <fn>
13 TRT01A Xanomeli… G60 TRUE tabulate… n n 152 0
14 TRT01A Xanomeli… G60 TRUE tabulate… p % 2.111 <fn>
15 TRT01A Xanomeli… G90 TRUE tabulate… n n 29 0
16 TRT01A Xanomeli… G90 TRUE tabulate… p % 0.403 <fn>
17 TRT01A Xanomeli… G110 TRUE tabulate… n n 0 0
18 TRT01A Xanomeli… G110 TRUE tabulate… p % 0 <fn>
19 TRT01A Xanomeli… GE120 TRUE tabulate… n n 0 0
20 TRT01A Xanomeli… GE120 TRUE tabulate… p % 0 <fn>
21 TRT01A Xanomeli… L60 TRUE tabulate… n n 0 0
22 TRT01A Xanomeli… L60 TRUE tabulate… p % 0 <fn>
23 TRT01A Xanomeli… G60 TRUE tabulate… n n 154 0
24 TRT01A Xanomeli… G60 TRUE tabulate… p % 1.638 <fn>
25 TRT01A Xanomeli… G90 TRUE tabulate… n n 29 0
26 TRT01A Xanomeli… G90 TRUE tabulate… p % 0.309 <fn>
27 TRT01A Xanomeli… G110 TRUE tabulate… n n 1 0
28 TRT01A Xanomeli… G110 TRUE tabulate… p % 0.011 <fn>
29 TRT01A Xanomeli… GE120 TRUE tabulate… n n 0 0
30 TRT01A Xanomeli… GE120 TRUE tabulate… p % 0 <fn>
31 <NA> TRT01A Placebo tabulate n n 86 0
32 <NA> TRT01A Placebo tabulate N N 254 0
33 <NA> TRT01A Placebo tabulate p % 0.339 <fn>
34 <NA> TRT01A Xanomeli… tabulate n n 72 0
35 <NA> TRT01A Xanomeli… tabulate N N 254 0
36 <NA> TRT01A Xanomeli… tabulate p % 0.283 <fn>
37 <NA> TRT01A Xanomeli… tabulate n n 96 0
38 <NA> TRT01A Xanomeli… tabulate N N 254 0
39 <NA> TRT01A Xanomeli… tabulate p % 0.378 <fn> Code
tbl <- tbl_ard_summary(
ard,
by = TRT01A,
# Add labels for each range
label = list(
L60 = "<60",
G60 = ">60",
G90 = ">90",
G110 = ">110",
GE120 = ">=120"
)
) |>
modify_header(
# Update label, add analysis value units
label ~ paste0("**Diastolic Blood Pressure (", data$VSORRESU[1], ")**"),
# Add N values to `by` variable labels
all_stat_cols() ~ "**{level}** \nN = {n}"
)
tbl