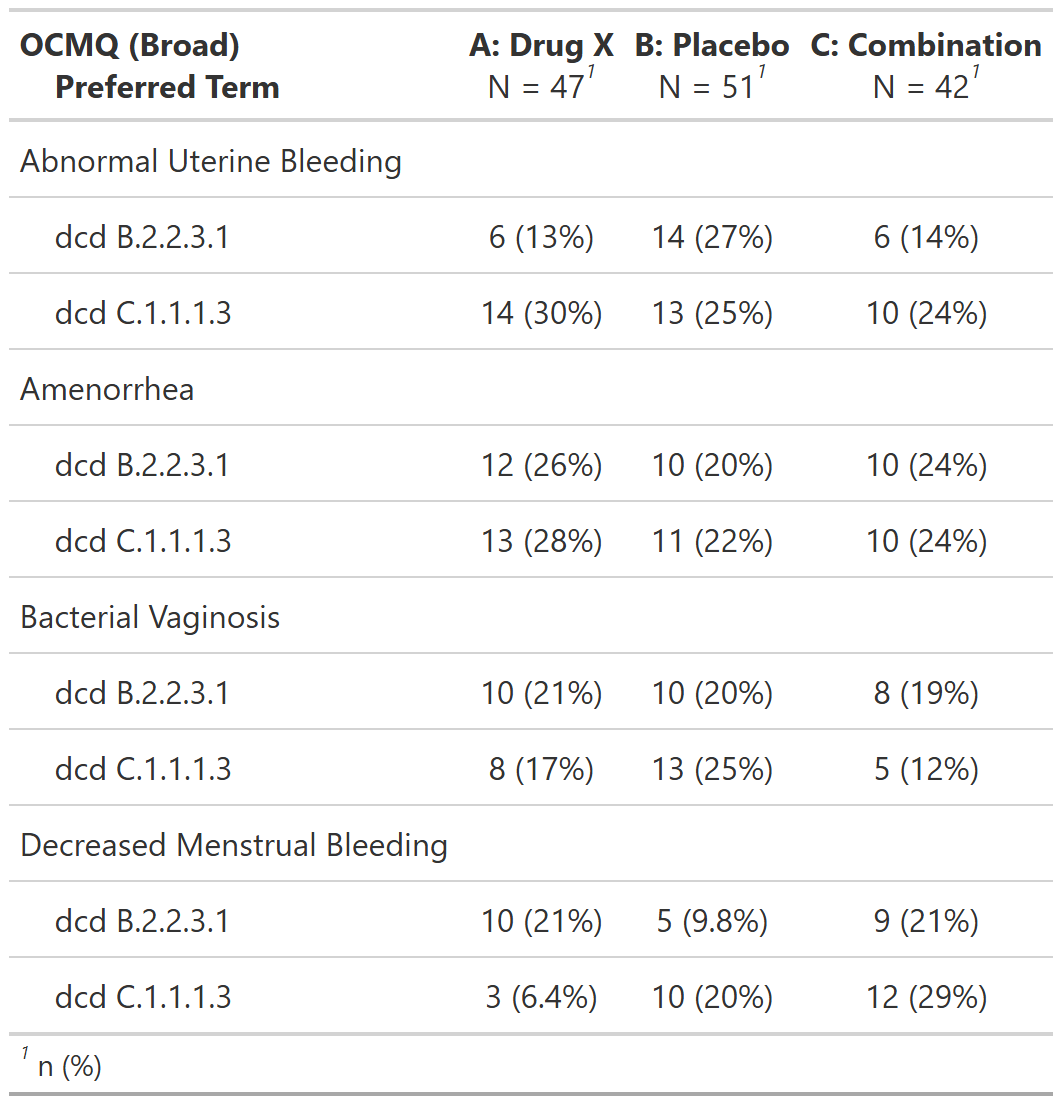
Subjects With Adverse Events by Female-Specific OCMQ (Broad) and Preferred Term, Female Safety Population, Pooled Analysis (or Trial X)
FDA Table 45
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adsl <- pharmaverseadam::adsl
adae <- pharmaverseadam::adae
set.seed(1)
adae <- adae |>
mutate(
OCMQ01SC = as.factor(sample(c("BROAD", "NARROW"), nrow(adae), replace = TRUE)),
OCMQ01NAM = if_else(
SEX == "F",
as.factor(sample(
c("Abnormal Uterine Bleeding", "Amenorrhea", "Bacterial Vaginosis", "Decreased Menstrual Bleeding"),
n(),
replace = TRUE
)),
NA_character_
)
)
# Pre-processing --------------------------------------------
data <- adae |>
filter(
SAFFL == "Y",
SEX == "F",
OCMQ01SC == "BROAD",
# Filtering to reduce the size of the demo table
AEDECOD %in% c("APPLICATION SITE IRRITATION", "ERYTHEMA", "APPLICATION SITE PRURITUS", "BACK PAIN")
) |>
select(OCMQ01SC, TRT01A, OCMQ01NAM, AEDECOD, USUBJID) |>
# setting an explicit level for NA values so empty strata combinations are shown.
mutate(across(everything(), ~ {
if (anyNA(.)) {
forcats::fct_na_value_to_level(as.factor(.), level = "<Missing>")
} else {
.
}
}))
# denominator values include only Female subjects in the arm with AEs
denom <- data |> distinct(USUBJID, TRT01A)
$tbl_hierarchical{cards} data frame: 126 x 15 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fun warning error gts_column
1 <NA> <NA> TRT01A Placebo tabulate n n 7 7 0 stat_1
2 <NA> <NA> TRT01A Placebo tabulate N N 40 40 0 stat_1
3 <NA> <NA> TRT01A Placebo tabulate p % 0.175 17.5 <fn> stat_1
4 <NA> <NA> TRT01A Xanomeli… tabulate n n 13 13 0 stat_2
5 <NA> <NA> TRT01A Xanomeli… tabulate N N 40 40 0 stat_2
6 <NA> <NA> TRT01A Xanomeli… tabulate p % 0.325 32.5 <fn> stat_2
7 <NA> <NA> TRT01A Xanomeli… tabulate n n 20 20 0 stat_3
8 <NA> <NA> TRT01A Xanomeli… tabulate N N 40 40 0 stat_3
9 <NA> <NA> TRT01A Xanomeli… tabulate p % 0.5 50.0 <fn> stat_3
10 TRT01A Placebo OCMQ01NAM Abnormal… AEDECOD APPLICAT… hierarch… n n 1 1 <fn> stat_1ℹ 116 more rowsℹ Use `print(n = ...)` to see more rows