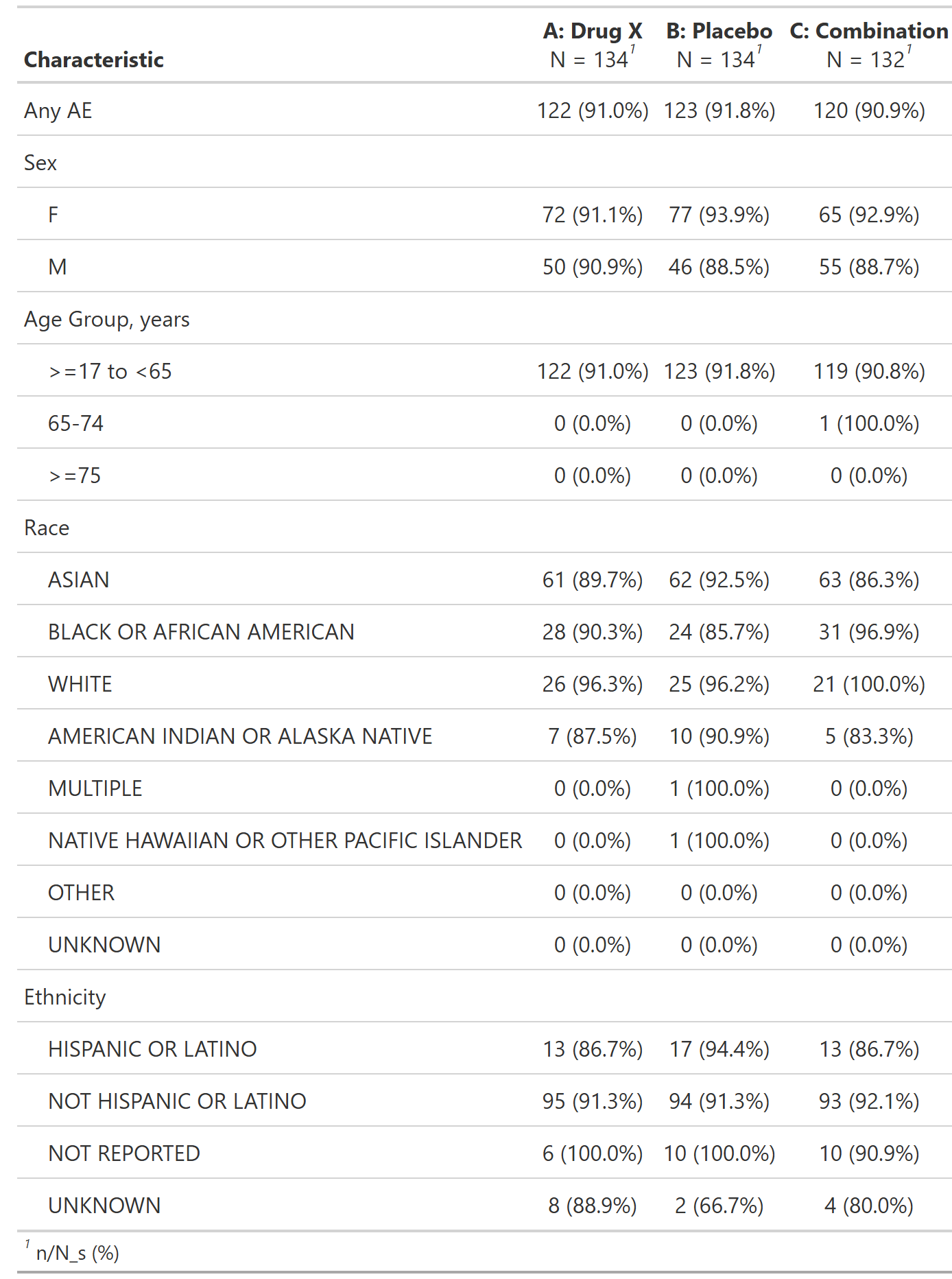
Overview of Adverse Events by Demographic Subgroup, Safety Population, Pooled Analysis (or Trial X)
FDA Table 51
table
FDA
safety
adverse events
Code
# Load libraries & data -------------------------------------
library(dplyr)
library(cards)
library(gtsummary)
adae <- pharmaverseadam::adae
adsl <- pharmaverseadam::adsl
adsl <- adsl |>
filter(SAFFL == "Y") |>
mutate(any_ae = "Any AE")
data <- adae |>
filter(
SAFFL == "Y",
TRTEMFL == "Y"
) |>
mutate(any_ae = "Any AE") |>
# subset to one AE per subject
dplyr::slice_tail(n = 1L, by = all_of(c("USUBJID", "ARM")))Code
# generate ARD for `n` statistic
ard_summary <- data |>
ard_categorical(
variables = c(any_ae, SEX, AGEGR1, RACE, ETHNIC),
by = ARM,
statistic = ~"n"
)
# generate ARD for `N_s` statistic
ard_Ns <- adsl |>
ard_categorical(
variables = c(any_ae, SEX, AGEGR1, RACE, ETHNIC),
by = ARM,
statistic = ~"n",
stat_label = ~ list(n = "N_s")
) |>
mutate(stat_name = "N_s")
# generate ARD for `p` statistic
ard_p <-
cards::bind_ard(
ard_summary,
ard_Ns
) |>
select(-stat_label) |>
tidyr::pivot_wider(names_from = stat_name, values_from = stat, values_fn = unlist) |>
rowwise() |>
mutate(
stat_name = "p",
stat_label = "%",
stat = if_else(n > 0, list(n / N_s), list(0)),
fmt_fun = list(label_round(digits = 1, scale = 100))
) |>
select(-n, -N_s)
# combine all ARDs
ard <- cards::bind_ard(ard_summary, ard_Ns, ard_p, ard_categorical(adsl, variables = ARM, statistic = ~"n")) |>
cards::tidy_ard_row_order()
# generate table from ARDs
tbl <-
tbl_ard_summary(
ard,
by = ARM,
# set variable labels
label = list(
SEX = "Sex",
AGEGR1 = "Age Group, years",
RACE = "Race",
ETHNIC = "Ethnicity"
)
) |>
gtsummary::modify_table_body(
\(x) x |> filter(!label %in% "any_ae")
) |>
gtsummary::modify_indent(columns = label, rows = variable == "any_ae", indent = 0L) |>
# update statistic footnote
gtsummary::modify_footnote_header(
footnote = "n/N_s (%)",
columns = all_stat_cols()
) |>
gtsummary::modify_header(all_stat_cols() ~ "**{level}** \n N = {n}")
tbl| Characteristic |
Placebo N = 861 |
Xanomeline High Dose N = 841 |
Xanomeline Low Dose N = 841 |
|---|---|---|---|
| Any AE | 65 (75.6%) | 75 (89.3%) | 77 (91.7%) |
| Sex | |||
| F | 40 (75.5%) | 36 (90.0%) | 44 (88.0%) |
| M | 25 (75.8%) | 39 (88.6%) | 33 (97.1%) |
| Age Group, years | |||
| >64 | 55 (76.4%) | 65 (89.0%) | 69 (90.8%) |
| 18-64 | 10 (71.4%) | 10 (90.9%) | 8 (100.0%) |
| Race | |||
| AMERICAN INDIAN OR ALASKA NATIVE | 0 (0.0%) | 1 (100.0%) | 0 (0.0%) |
| BLACK OR AFRICAN AMERICAN | 6 (75.0%) | 7 (77.8%) | 4 (66.7%) |
| WHITE | 59 (75.6%) | 67 (90.5%) | 73 (93.6%) |
| Ethnicity | |||
| HISPANIC OR LATINO | 3 (100.0%) | 3 (100.0%) | 6 (100.0%) |
| NOT HISPANIC OR LATINO | 62 (74.7%) | 72 (88.9%) | 71 (91.0%) |
| 1 n/N_s (%) | |||

$tbl_ard_summary{cards} data frame: 103 x 12 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 <NA> any_ae attribut… label Variable… any_ae <fn> <NA>
2 <NA> any_ae attribut… class Variable… logical NULL <NA>
3 ARM Placebo any_ae Any AE categori… n n 65 0 stat_1
4 ARM Placebo SEX F categori… n n 40 0 stat_1
5 ARM Placebo SEX M categori… n n 25 0 stat_1
6 ARM Placebo AGEGR1 >64 categori… n n 55 0 stat_1
7 ARM Placebo AGEGR1 18-64 categori… n n 10 0 stat_1
8 ARM Placebo RACE AMERICAN… categori… n n 0 0 stat_1
9 ARM Placebo RACE BLACK OR… categori… n n 6 0 stat_1
10 ARM Placebo RACE WHITE categori… n n 59 0 stat_1
11 ARM Placebo ETHNIC HISPANIC… categori… n n 3 0 stat_1
12 ARM Placebo ETHNIC NOT HISP… categori… n n 62 0 stat_1
13 ARM Placebo any_ae Any AE categori… N_s N_s 86 0 stat_1
14 ARM Placebo SEX F categori… N_s N_s 53 0 stat_1
15 ARM Placebo SEX M categori… N_s N_s 33 0 stat_1
16 ARM Placebo AGEGR1 >64 categori… N_s N_s 72 0 stat_1
17 ARM Placebo AGEGR1 18-64 categori… N_s N_s 14 0 stat_1
18 ARM Placebo RACE AMERICAN… categori… N_s N_s 0 0 stat_1
19 ARM Placebo RACE BLACK OR… categori… N_s N_s 8 0 stat_1
20 ARM Placebo RACE WHITE categori… N_s N_s 78 0 stat_1
21 ARM Placebo ETHNIC HISPANIC… categori… N_s N_s 3 0 stat_1
22 ARM Placebo ETHNIC NOT HISP… categori… N_s N_s 83 0 stat_1
23 ARM Placebo any_ae Any AE categori… p % 0.756 <fn> stat_1
24 ARM Placebo SEX F categori… p % 0.755 <fn> stat_1
25 ARM Placebo SEX M categori… p % 0.758 <fn> stat_1
26 ARM Placebo AGEGR1 >64 categori… p % 0.764 <fn> stat_1
27 ARM Placebo AGEGR1 18-64 categori… p % 0.714 <fn> stat_1
28 ARM Placebo RACE AMERICAN… categori… p % 0 <fn> stat_1
29 ARM Placebo RACE BLACK OR… categori… p % 0.75 <fn> stat_1
30 ARM Placebo RACE WHITE categori… p % 0.756 <fn> stat_1
31 ARM Placebo ETHNIC HISPANIC… categori… p % 1 <fn> stat_1
32 ARM Placebo ETHNIC NOT HISP… categori… p % 0.747 <fn> stat_1
33 ARM Xanomeli… any_ae Any AE categori… n n 75 0 stat_2
34 ARM Xanomeli… SEX F categori… n n 36 0 stat_2
35 ARM Xanomeli… SEX M categori… n n 39 0 stat_2
36 ARM Xanomeli… AGEGR1 >64 categori… n n 65 0 stat_2
37 ARM Xanomeli… AGEGR1 18-64 categori… n n 10 0 stat_2
38 ARM Xanomeli… RACE AMERICAN… categori… n n 1 0 stat_2
39 ARM Xanomeli… RACE BLACK OR… categori… n n 7 0 stat_2
40 ARM Xanomeli… RACE WHITE categori… n n 67 0 stat_2ℹ 63 more rowsℹ Use `print(n = ...)` to see more rows