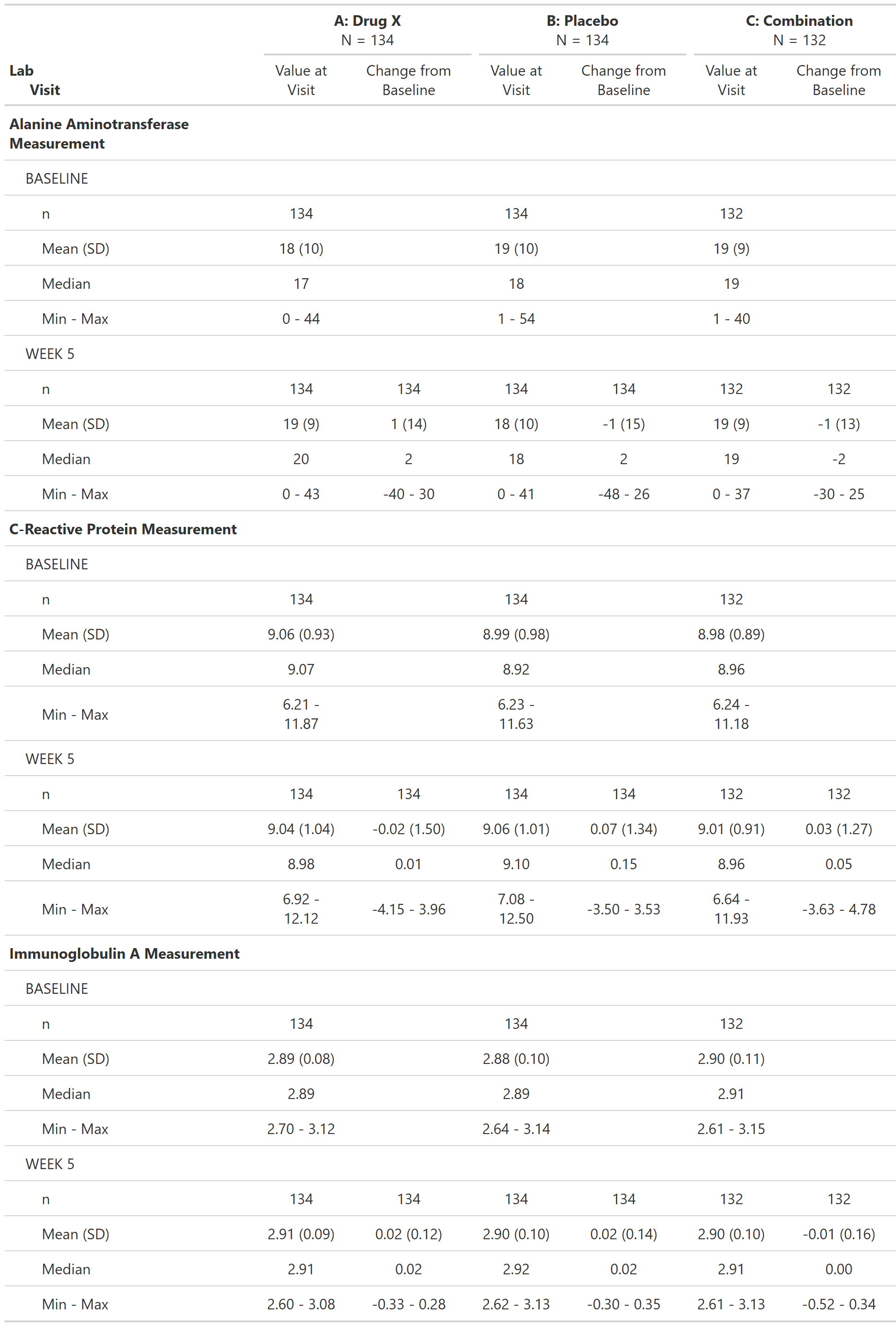
Laboratory Test Results and Change from Baseline by Visit
Roche LBT01
table
Roche
parallel-group
change from baseline
Code
# Load libraries & data -------------------------------------
library(gtsummary)
library(dplyr)
library(crane)
adsl <- pharmaverseadam::adsl
adlb <- pharmaverseadam::adlb
# Pre-processing --------------------------------------------
data <- adlb |>
dplyr::filter(
SAFFL == "Y", # safety population
PARAMCD %in% c("ALT", "BILI", "CREAT")
# This step can be used to filter for valid visits. Since the table creation
# code specifies the visits, we can omit that step here.
)Code
tbl <- data |>
filter(
AVISIT %in% c("Baseline", "Week 4") # specify visits to display in table
) |>
# apply tbl_baseline_chg function to each level of PARAM in the data
gtsummary::tbl_strata_nested_stack(
strata = PARAM,
~ .x |>
crane::tbl_baseline_chg(
baseline_level = "Baseline",
by = "TRT01A",
# this dataset has a treatment level called "Screen Failure".
# Filtering this out as adsl is used to calculate the denominator.
denominator = adsl |> filter(TRT01A != "Screen Failure")
)
) |>
# bold lab test labels
gtsummary::modify_bold(columns = label, rows = tbl_indent_id1 > 0L) |>
# update header labels
gtsummary::modify_header(label = "**Lab \n\U00A0\U00A0\U00A0\U00A0 Visit**") |>
gtsummary::modify_spanning_header(gtsummary::all_stat_cols() ~ "**{level}** \nN = {n}")
tbl
$`PARAM="Alanine Aminotransferase (U/L)"`
$`PARAM="Alanine Aminotransferase (U/L)"`$tbl_baseline_chg{cards} data frame: 108 x 12 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 TRT01A Placebo AVAL Baseline summary mean Mean 17.57 <fn> stat_1
2 TRT01A Placebo AVAL Baseline summary sd SD 9.216 <fn> stat_1
3 TRT01A Placebo AVAL Baseline summary median Median 15 <fn> stat_1
4 TRT01A Placebo AVAL Baseline summary min Min 7 <fn> stat_1
5 TRT01A Placebo AVAL Baseline summary max Max 69 <fn> stat_1
6 TRT01A Placebo AVAL Week 4 summary mean Mean 18.658 <fn> stat_1
7 TRT01A Placebo AVAL Week 4 summary sd SD 12.908 <fn> stat_1
8 TRT01A Placebo AVAL Week 4 summary median Median 16 <fn> stat_1
9 TRT01A Placebo AVAL Week 4 summary min Min 6 <fn> stat_1
10 TRT01A Placebo AVAL Week 4 summary max Max 107 <fn> stat_1ℹ 98 more rowsℹ Use `print(n = ...)` to see more rows
$`PARAM="Bilirubin (umol/L)"`
$`PARAM="Bilirubin (umol/L)"`$tbl_baseline_chg{cards} data frame: 108 x 12 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 TRT01A Placebo AVAL Baseline summary mean Mean 9.703 <fn> stat_1
2 TRT01A Placebo AVAL Baseline summary sd SD 3.965 <fn> stat_1
3 TRT01A Placebo AVAL Baseline summary median Median 8.55 <fn> stat_1
4 TRT01A Placebo AVAL Baseline summary min Min 5.13 <fn> stat_1
5 TRT01A Placebo AVAL Baseline summary max Max 25.65 <fn> stat_1
6 TRT01A Placebo AVAL Week 4 summary mean Mean 11.126 <fn> stat_1
7 TRT01A Placebo AVAL Week 4 summary sd SD 13.57 <fn> stat_1
8 TRT01A Placebo AVAL Week 4 summary median Median 8.55 <fn> stat_1
9 TRT01A Placebo AVAL Week 4 summary min Min 5.13 <fn> stat_1
10 TRT01A Placebo AVAL Week 4 summary max Max 124.83 <fn> stat_1ℹ 98 more rows
ℹ Use `print(n = ...)` to see more rows
$`PARAM="Creatinine (umol/L)"`
$`PARAM="Creatinine (umol/L)"`$tbl_baseline_chg{cards} data frame: 108 x 12 group1 group1_level variable variable_level context stat_name stat_label stat fmt_fun warning error gts_column
1 TRT01A Placebo AVAL Baseline summary mean Mean 97.651 <fn> stat_1
2 TRT01A Placebo AVAL Baseline summary sd SD 17.779 <fn> stat_1
3 TRT01A Placebo AVAL Baseline summary median Median 97.24 <fn> stat_1
4 TRT01A Placebo AVAL Baseline summary min Min 61.88 <fn> stat_1
5 TRT01A Placebo AVAL Baseline summary max Max 159.12 <fn> stat_1
6 TRT01A Placebo AVAL Week 4 summary mean Mean 98.641 <fn> stat_1
7 TRT01A Placebo AVAL Week 4 summary sd SD 18.557 <fn> stat_1
8 TRT01A Placebo AVAL Week 4 summary median Median 97.24 <fn> stat_1
9 TRT01A Placebo AVAL Week 4 summary min Min 70.72 <fn> stat_1
10 TRT01A Placebo AVAL Week 4 summary max Max 176.8 <fn> stat_1ℹ 98 more rows
ℹ Use `print(n = ...)` to see more rows