Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionAndré Veríssimo
Tymoteusz Makowski
Pedro Silva
Vedha Viyash
APPSILON
February 1, 2024
The R Consortium Submission Working Group has now successfully made two pilot submissions to the FDA. All the submissions done by the group are focused on improving practices for R-based clinical trial regulatory submissions. Now, the R submission Working Groups, in collaboration with Appsilon and Posit, are exploring new technologies such as Containers and WebAssembly. In this article, we dive into the details of this exploration.
This pilot was initially submitted on November 22, 2021. This submission was the first publicly available R-based submission to the FDA. This was a test submission that aimed to explore the submission of an R package to the FDA following the eCTD specifications. The submission included an R package, R scripts for analysis, R-based analysis data reviewed guide (ADRG), and other important components. The final response letter from the FDA was received on March 14, 2022.
This was one of the first submission packages containing a Shiny application. The main goal of this pilot was to test the submission of an R-based Shiny application bundled into a submission package and transfer it successfully to FDA reviewers. The submitted application was built using the datasets and analyses that were used for the R Submission Pilot 1. The deployed version of this application is available on this site. Alternatively, a Rhino-based version of the application can be found here.
The final response letter from the FDA was reviewed on September 27, 2023.

In this submission, there were many open-source R packages that were used to create and execute the Shiny application. A very well-known shiny-based interactive exploration framework {teal} was used mainly for analyzing the clinical trial data; this package is included in the pharmaverse package repository. The full list of open-source and proprietary R analysis packages is available on this Analysis Data Reviewer’s Guide prepared by the R Consortium R Submissions Working Group for the Pilot 2.
This pilot was successfully submitted to the FDA on Aug 28, 2023. This was the first publicly available R submission that included R scripts to produce ADaM datasets and TLFs. Both the ADaMs (SDTM .xpt sources from the CDISC Pilot study) and the TLFs (ADaMs .xpt sourced from the ADaMs generated in R by the Pilot 3 team) were created using R. The next step for this pilot is to await FDA’s review and approval, which may take several months to complete.
This pilot aims to explore using technologies such as containers and WebAssembly software to package a Shiny application into a self-contained unit, streamlining the transfer and execution process for enhanced efficiency.
This pilot is expected to be divided into two parallel submissions:
(a) will investigate WebAssembly and
(b) will investigate containers.
Our team at Appsilon teamed up with the dynamic Pilot 4 crew to explore WebAssembly technology and containers. George Stagg and Winston Chang also joined the working group to discuss the web-assembly portion of Pilot 4. This partnership brought together our engineering prowess to contribute to these tools, injecting fresh perspectives into the ongoing pilot project.
Some of the outcomes of the collaboration:
We were able to set up a robust container environment for this pilot project.
We aided the progress made on the use of both experimental technologies: containers and WebAssembly.
We developed a working prototype submission using Podman container technology.
We developed a working early-stage prototype for wrapping a small Shiny application using WebAssembly.

WebAssembly allows languages like R to be executed at near-native speed directly within web browsers, providing users with the ability to run R code without having R installed locally. WebR is essentially the R programming language adapted to run in a web browser environment using WebAssembly. This project is under active development.
The deployed example of the Shiny app running on webR is available here. Check out the video of the application running below.
During this pilot, engineers at Appsilon developed a prototype of a Shiny application running on webR. The application reuses most of the code from the previous pilot apps with some tweaks and a couple of hacks/changes to get around non CRAN dependencies, specially for data loading, WebR compatibilities, and shimming some of the functionality from {teal} and other packages that are (for now) not available on CRAN.
During the second iteration, which was recently held, Pedro Silva shared the process of developing this Shiny app running on webR.
Leverage the last 2 iterations of the application
Reuse as much code as possible
Avoid touching the logic part
Restrict the number of dependencies to packages on CRAN
Here is the list of dependencies to packages on CRAN; those that worked are colored green, and those that were removed are marked in orange. We ended up with just 3 problematic dependencies (bold).
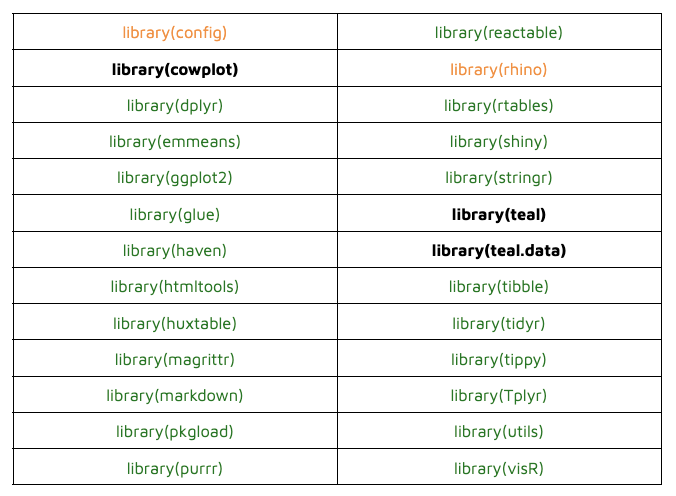
Issues with library(cowplot):
Solution:
Issues with library(teal):
Solution:
Redo the UI
Load modules directly
Recreate filter functionality
Issues with library(teal.data):
Solution:
Leverage shinylive and httpuv to export and serve the application
Shinylive can help streamline the export process
Problems
shiny.live won’t let us have non-R files in the application directory - this is an outstanding bug that George asked us to raise an issue for.
We wouldn’t be able to run the application as a traditional shiny app.
Solution:
{httpuv} can help serve the application
The figure below shows an overview of what we ended with:
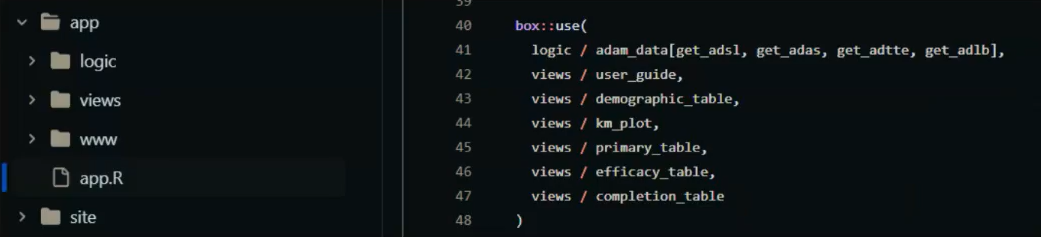
Some of the issues and solutions found at the very beginning:
The FDA was previously told that the shiny application being prepared for the Pilot 4 submission would not be a 1 to 1 mapping from the previous one submitted for the Pilot 2 due to certain constraints such as {teal} not being on CRAN; however, this didn’t represent a problem for them since they would mainly like to test the technology.
Pedro Silva, one of the engineers working on the development of this app, mentioned “While WebR is still in development, it shows tremendous promise! The loading is definitely still a pain point (over 100mb to set up the environment!) but it will only get better moving forward.”

Containerization, particularly through technologies like Docker, Podman or Singularity, offers several advantages for deploying Shiny apps.
Choosing the right container was a question that arose in this project. Although Docker is the most popular, we decided to move forward with Podman.
In our exploration of containerization tools for deploying Shiny applications, we’ve identified key distinctions between Docker and Podman that influenced our choice.
Podman stands out for its daemonless architecture, enhancing security by eliminating the need for a central daemon process. Unlike Docker, Podman supports running containers as non-root users, a critical feature for meeting FDA reviewer requirements. Developed by Red Hat and maintained as an open-source project, Podman prioritizes security with its rootless container support, offering a robust solution for security-conscious users.
A Container-based method to deploy Pilot 2 Shiny App.
Configurable Podman Dockerfile / docker-compose.yml
Documentation on creating the container
CI: Automated build on amd64 and arm64 platforms
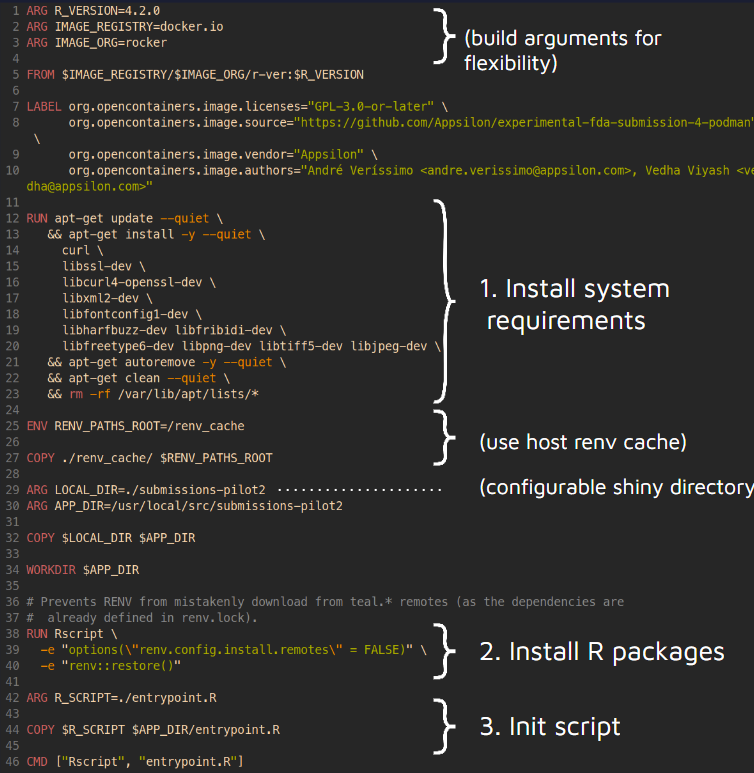
Below is the dockerfile (recipe) for the container:
The next steps are waiting for the review of Pilot 3 by the FDA and to submit the two sections that explore the new technologies to regulatory authorities. Thanks to the collaboration between the R Submission Working Group and other institutions, there is already a working prototype of a {teal}-like Shiny application running on webR and further exploration with Podman is underway.

Appsilon is working on the side with Rhino compatibility; eventually, we might be able to just add this framework into the Pilot 4 application.
{teal}
This and other packages might be on CRAN soon. We could incorporate them after that, replacing the shims created for this version.
Boot Time
We need to improve boot time (remove dependencies and keep working on webR).
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, union2026-02-20 17:01:19.114928
@online{veríssimo2024,
author = {Veríssimo, André and Makowski, Tymoteusz and Silva, Pedro
and Viyash, Vedha and , APPSILON},
title = {Testing {Containers} and {WebAssembly} in {Submissions} to
the {FDA}},
date = {2024-02-01},
url = {https://pharmaverse.github.io/blog/posts/2024-02-01_containers_webassembly_submission/containers_and_webassembly_submissions.html},
langid = {en}
}