Disclaimer: This blog contains opinions that are of the authors alone and do not necessarily reflect the strategy of their respective organizations.
We are pleased to introduce a new addition to the pharmaverse: {docorator}, an R package developed by GSK to facilitate the creation of production displays within pharma workflows. If you’ve ever spent hours perfecting a table or figure, only to wrestle with adding compliant headers, footers, and perfect margins for a polished product, {docorator} might be for you.
For full documentation, visit the website here: https://gsk-biostatistics.github.io/docorator/
What is docorator?
The package name, {docorator}, signifies its core functionality: decorate and output. The package takes TLGs generated by popular R packages such as {gt} (and by extension {tfrmt} and {gtsummary}) and {ggplot2} and “decorates” them with custom headers, footers, etc. in production-ready outputs.
The {docorator} package is built on a foundation of reproducibility and flexibility through its multi-rendering framework. While the primary and most robust rendering engine is PDF (leveraging Rmarkdown/Quarto and LaTeX internally), basic RTF support is also available, with the underlying framework designed for expansion to other output formats in the future.
How docorator complements the pharmaverse
This package can be considered complementary to the pharmaverse packages {clinify} and {gridify}. It offers an additional option for R users seeking to meet organizational requirements for specific output types, as well as standardized headers, footers, margins, and sizing. A key feature of {docorator} is its PDF support with embedded table values. By rendering to PDF via RMarkdown/Quarto and LaTeX, the displayed values become embedded within the PDF document, which enhance traceability. For {gt} tables specifically, automated table scaling (i.e. column resizing) is applied by default to assist with fitting the display to the available space.
Example: PDF render
The process of rendering a display involves two main steps:
Create a
docoratorobject: This step uses theas_docorator()function to add headers, footers, and other styling elements.Render an output: A simple function call generates the final document.
Below is an illustrative example:
# This code assumes a display object called `t_demog` has been created
t_demog |>
as_docorator(
display_name = "t_demog",
display_loc = "path/to/output/folder",
header = fancyhead(
fancyrow(left = "Study ABC123", center = NA, right = doc_pagenum()),
fancyrow(left = "Safety Population", center = NA, right = NA),
fancyrow(left = NA, center = "Demographic Summary", right = NA)
),
footer = fancyfoot(
fancyrow(left = "path/to/t_demog.R", right = doc_datetime())
)
) |>
render_pdf()Note that headers and footers are specified via the header and footer arguments using the fancyhead() and fancyfoot() functions, respectively. These functions, inspired by (and for PDF, powered by) the fancyhdr LaTeX library, allow users to flexibly add multiple rows of headers and/or footers with left, center, and/or right aligned text. Rows of headers/footers are added via the fancyrow() function which has arguments for where the custom text should be added.
Here’s the result:
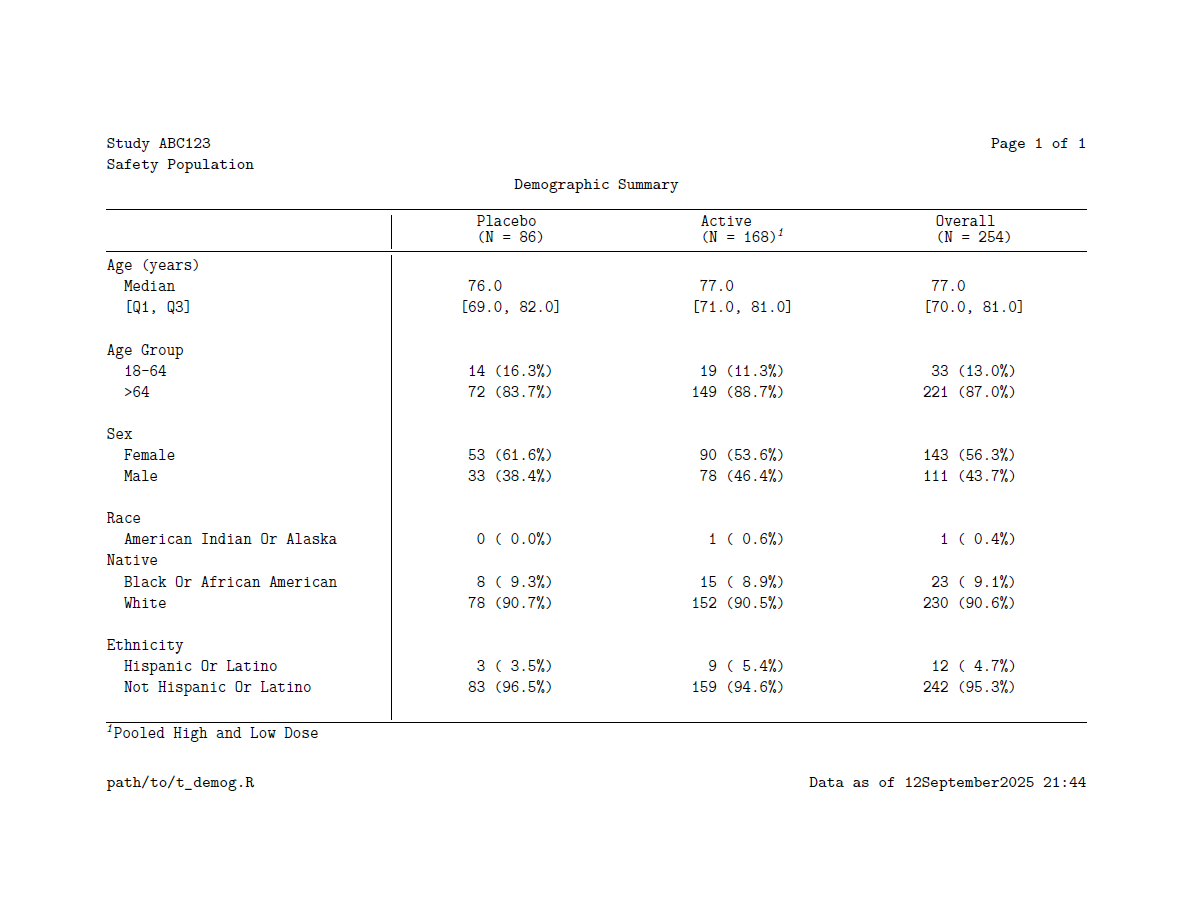 .
.
Example: multiple renders
In pharma, it’s very common for displays to be utilized across multiple reports, presentations, and various downstream systems. {docorator} is engineered for reuse, offering several powerful features:
- Multi-render capability: Users can render to multiple output types using pipeable syntax.
# Continuing from the previous example:
t_demog |>
as_docorator(
display_name = "t_demog",
display_loc = "path/to/output/folder",
header = fancyhead(
fancyrow(left = "Study ABC123", center = NA, right = doc_pagenum()),
fancyrow(left = "Safety Population", center = NA, right = NA),
fancyrow(left = NA, center = "Demographic Summary", right = NA)
),
footer = fancyfoot(
fancyrow(left = "path/to/t_demog.R", right = doc_datetime())
)
) |>
render_pdf() |>
render_rtf() # This will generate both PDF and RTF outputs- The
docoratorobject artifact:
Executing as_docorator() does not immediately render your display. Instead, it creates a docorator object that stores all relevant display information in an .rds file. This object serves as an artifact for future rendering or other downstream requirements. For the above example, t_demog.rds will be saved in the folder “path/to/output/folder” alongside the “t_demog.pdf” and “t_demog.rtf” output documents.
A note on LaTeX dependencies
As noted in its README, {docorator} requires a LaTeX installation for PDF rendering. If using {tinytex}, the TinyTeX bundle is recommended, which can be installed via the following code:
tinytex::install_tinytex(bundle = "TinyTeX")Get in touch!
We encourage you to try {docorator} and provide any suggestions or report problems via the GitHub issues.
Reuse
Citation
@online{krouse2025,
author = {Krouse, Becca},
title = {Introducing Docorator to the Pharmaverse},
date = {2025-12-19},
url = {https://pharmaverse.github.io/blog/posts/2025-12-19_introducing/introducing_docorator.html},
langid = {en}
}