Overview
In the pharmaceutical industry, and many other fields that rely heavily on data reporting, there is often a need to create figures and tables with specific graphical arrangements. These could be titles, subtitles, captions, footnotes, and other text elements that provide important context to the data being shown.
However, creating the headers and footers etc. and correctly positioning them around the output can be challenging, often requiring fine-tuning. This can be time-consuming and can lead to inconsistencies in the way the figures and tables are presented across different projects.
gridify builds on the base R grid package and makes it easy to add flexible and customizable information around a figure or table using a pre-defined or custom layout. The gridify package works with all of the following input types, creating consistency when using various different inputs:
grob, gtable, ggplot, flextable, gt, base R plots (by formula)
Whilst rtables are not directly supported, we can use rtables with gridify by first converting them to flextable (with rtables.officer).
As gridify is based on the graphical tool grid, any figure or table inputs are converted to a grob object in gridify and the result of using gridify is always a graphical image.
Installation
You can install the newest release version from CRAN:
install.packages("gridify")Or you can install the newest development version from Pharmaverse GitHub (example):
# install.packages("remotes")
remotes::install_github("pharmaverse/gridify", build_manual = TRUE)Example
The workflow of the package is as follows:
- Create your object (
ggplot,gtetc.) - Choose a layout (predefined or custom). Use
get_layouts()to see the predefined options - Use
gridify()to create agridifyobject - Print the
gridifyobject to see empty cells - Use
set_cell()to fill in the various text elements in the layout (headers, footers etc.)
The following example uses a table created by the gt package and the gridify layout pharma_layout_base().
library(gridify)
# install.packages("gt")
# gt needs gtable
# install.packages("gtable")
library(gt)
# (to use |> version 4.1.0 of R is required, for lower versions we recommend %>% from magrittr)
tab <- gt::gt(head(mtcars, n = 10)) |>
gt::tab_options(
table.width = gt::pct(100),
data_row.padding = gt::px(10),
table_body.hlines.color = "white",
# gt font size is in pixels
# Multiply points by 96/72 to get pixels
table.font.size = 10 * 96 / 72,
table.font.names = "sans"
)
# Use `gridify()` to create a `gridify` object
gridify_object <- gridify(
object = tab,
# Choose a layout (predefined or custom)
layout = pharma_layout_base(
margin = grid::unit(c(0.5, 0.5, 0.5, 0.5), "inches"),
global_gpar = grid::gpar(fontfamily = "sans", fontsize = 10)
)
)
# Print the `gridify` object to see empty cells
gridify_object
# Use `set_cell()` to fill in the various text elements in the layout (headers etc.)
gridify_object_fill <- gridify_object |>
set_cell("header_left_1", "My Company") |>
set_cell("header_left_2", "<PROJECT> / <INDICATION>") |>
set_cell("header_left_3", "<STUDY>") |>
set_cell("header_right_1", "CONFIDENTIAL") |>
set_cell("header_right_2", "<Draft or Final>") |>
set_cell("header_right_3", "Data Cut-off: YYYY-MM-DD") |>
set_cell("output_num", "<Table> xx.xx.xx") |>
set_cell("title_1", "<Title 1>") |>
set_cell("title_2", "<Title 2>") |>
set_cell("title_3", "<Optional Title 3>") |>
set_cell("by_line", "By: <GROUP>, <optionally: Demographic parameters>") |>
set_cell("note", "<Note or Footnotes>") |>
set_cell("references", "<References:>") |>
set_cell("footer_left", "Program: <PROGRAM NAME>, YYYY-MM-DD at HH:MM") |>
set_cell("footer_right", "Page xx of nn") |>
set_cell("watermark", "DRAFT")
gridify_object_fill
print(gridify_object_fill)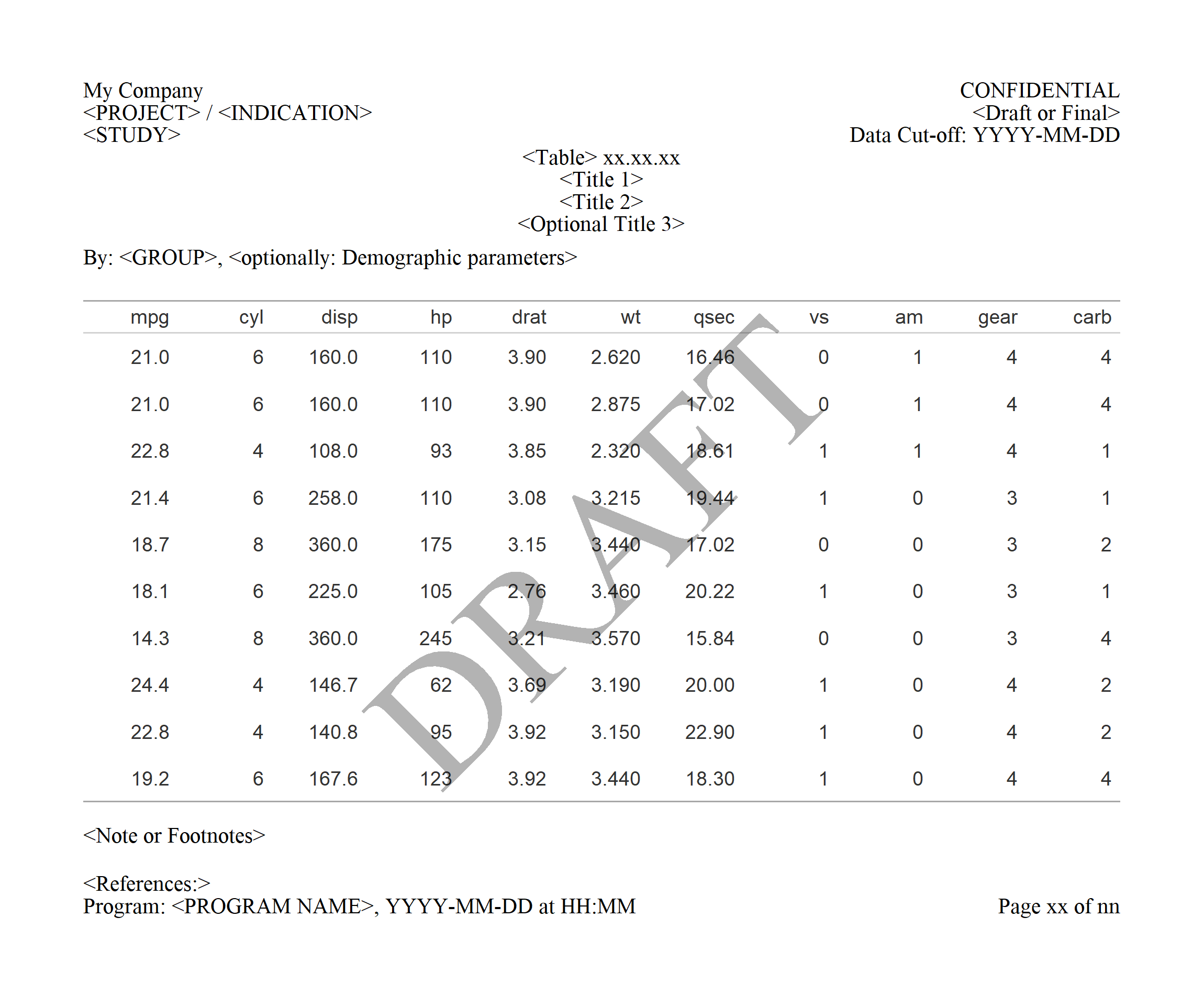
Note: Get the image usingexport_to(gridify_object_fill, to = "mypng.png", res = 300, width = 2300, height = 1900)
Documentation Guide
For more information please visit the following vignettes:
-
Getting Started
vignette("gridify", package = "gridify")- A case study in how the above example is constructed. -
Simple Examples
vignette("simple_examples", package = "gridify")- A showcase of implementations ofgridifyfor the various possible inputs. -
Multi-Page Examples
vignette("multi_page_examples", package = "gridify")- Showing how to usegridifyin more complex situations e.g a for-loop for multiple results. -
Create Custom Layout
vignette("create_custom_layout", package = "gridify")- An explanation on how to create a custom layout to use ingridify. -
Transparency of gridify
vignette("transparency", package = "gridify")- How to extract the raw grid code to reproduce agridifyobject.
Related packages
Other packages exist which add headers, footers, and other elements to figures and tables; most of the input classes to gridify already support these features. However, gridify was created not to supersede these, but to be used in conjunction with, in a way that is flexible for all use cases and consistent across various inputs.