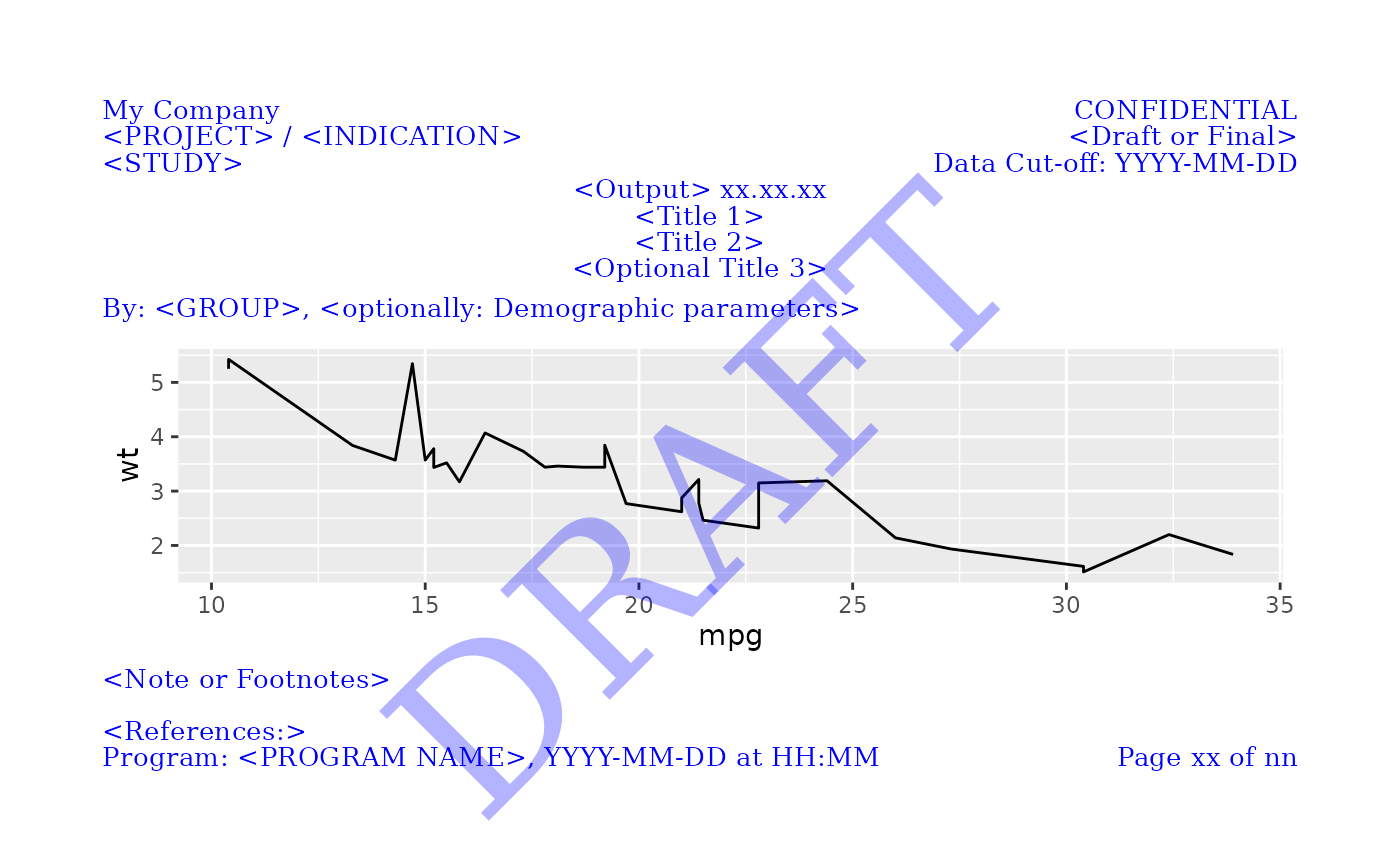
This function sets up the general structure for positioning text elements for pharma layouts. It defines the layout with specified margins, global graphical parameters, and height adjustment. The layout includes cells for headers, titles, footers, and optional elements like watermarks.
Arguments
- margin
A
grid::unitobject defining the margins of the layout (top, right, bottom, left) in inches. Default isgrid::unit(c(1, 1, 1, 1), "inches").- global_gpar
A list specifying global graphical parameters to change in the layout. Default is NULL, however the defaults in the layout are:
fontfamily = "Serif", fontsize = 9 and lineheight = 0.95, which can be overwritten alongside other graphical parameters found bygrid::get.gpar().- background
A string specifying the background fill colour. Default
grid::get.gpar()$fillfor a white background.- adjust_height
A logical value indicating whether to adjust the height of the layout. Default is
TRUE.
Details
This function is primarily used internally by other layout functions such as pharma_layout_A4() and
pharma_layout_letter() to create specific layouts.
Note
The Font Issue Information:
Changes to the fontfamily may be ignored by some devices, but is supported by PostScript, PDF, X11, Windows, and Quartz. The fontfamily may be used to specify one of the Hershey Font families (e.g., HersheySerif, serif), and this specification will be honoured on all devices.
If you encounter this warning, you can register the fonts using the extrafont package:
If you still see the warning while using RStudio, try changing the graphics backend.
Negative Dimensions Issues:
grobs from the grid package and ggplot2 objects (when converted to grobs by gridify) may appear distorted in the output if there is insufficient space in the window, caused by negative dimensions. This should be resolved. However, if this is affecting your layout, please increase your window size or only use static heights/widths for custom layouts.
The negative dimensions are caused by the way grid handles null and npc heights/widths so if some dimensions are
static, then the npc or null values may cause unexpected behaviour when the window size is too small.
It was resolved by setting a minimum size of the object in the gridify object to 1 inch for each dimension.
The following example demonstrates this behaviour Try resizing your window:
library(grid)
library(ggplot2)
grid.newpage()
object <- ggplot2::ggplotGrob(ggplot(mtcars, aes(mpg, wt)) + geom_line())
grid::grid.draw(
grid::grobTree(
grid::grobTree(
grid::editGrob(
object,
vp = grid::viewport(
# height = grid::unit.pmax(grid::unit(1, "npc"), grid::unit(1, "inch")),
# width = grid::unit.pmax(grid::unit(1, "npc"), grid::unit(1, "inch"))
)
),
vp = grid::viewport(
layout.pos.row = 2,
layout.pos.col = 1:3
)
),
vp = grid::viewport(
layout = grid::grid.layout(
nrow = 3,
ncol = 3,
heights = grid::unit(c(9, 1, 9), c("cm", "null", "cm"))
)
)
)
)gt Font Size Issue:
When specifying font sizes, the gt package interprets values as having the
unit pixels (px), whilst the grid package, on which gridify is built,
assumes points (pt). As a result, even if you set the font sizes in both
gt and gridify (using grid::gpar()) to the same number, they may
still appear different. To convert point size to pixel size, multiply the point size by 96 / 72.
Examples
# Create a general pharma layout with default settings
pharma_layout_base()
#> gridifyLayout object
#> ---------------------
#> Layout dimensions:
#> Number of rows: 14
#> Number of columns: 3
#>
#> Heights of rows:
#> Row 1: 1 lines
#> Row 2: 1 lines
#> Row 3: 1 lines
#> Row 4: 1 lines
#> Row 5: 1 lines
#> Row 6: 1 lines
#> Row 7: 0 lines
#> Row 8: 0 lines
#> Row 9: 1 lines
#> Row 10: 1 null
#> Row 11: 1 lines
#> Row 12: 1 lines
#> Row 13: 0 lines
#> Row 14: 1 lines
#>
#> Widths of columns:
#> Column 1: 0.333333333333333 npc
#> Column 2: 0.333333333333333 npc
#> Column 3: 0.333333333333333 npc
#>
#> Object Position:
#> Row: 10
#> Col: 1-3
#> Width: 1
#> Height: 1
#>
#> Object Row Heights:
#> Row 10: 1 null
#>
#> Margin:
#> Top: 1 inches
#> Right: 1 inches
#> Bottom: 1 inches
#> Left: 1 inches
#>
#> Global graphical parameters:
#> fill: white
#> col: black
#> lty: solid
#> lwd: 1
#> cex: 1
#> fontsize: 9
#> lineheight: 0.95
#> font: 1
#> fontfamily: serif
#> alpha: 1
#> lineend: round
#> linejoin: round
#> linemitre: 10
#> lex: 1
#>
#> Background colour:
#> white
#>
#> Default Cell Info:
#> header_left_1:
#> row:1, col:1, text:NULL, mch:Inf, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> header_left_2:
#> row:2, col:1, text:NULL, mch:Inf, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> header_left_3:
#> row:3, col:1, text:NULL, mch:Inf, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> header_right_1:
#> row:1, col:3, text:CONFIDENTI..., mch:Inf, x:1, y:0.5, hjust:1, vjust:0.5, rot:0,
#> header_right_2:
#> row:2, col:3, text:NULL, mch:Inf, x:1, y:0.5, hjust:1, vjust:0.5, rot:0,
#> header_right_3:
#> row:3, col:3, text:NULL, mch:Inf, x:1, y:0.5, hjust:1, vjust:0.5, rot:0,
#> output_num:
#> row:4, col:2, text:NULL, mch:Inf, x:0.5, y:0.5, hjust:0.5, vjust:0.5, rot:0,
#> title_1:
#> row:5, col:2, text:NULL, mch:Inf, x:0.5, y:0.5, hjust:0.5, vjust:0.5, rot:0,
#> title_2:
#> row:6, col:2, text:NULL, mch:Inf, x:0.5, y:0.5, hjust:0.5, vjust:0.5, rot:0,
#> title_3:
#> row:7, col:2, text:NULL, mch:Inf, x:0.5, y:0.5, hjust:0.5, vjust:0.5, rot:0,
#> by_line:
#> row:8-9, col:1-3, text:NULL, mch:Inf, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> note:
#> row:11, col:1-3, text:NULL, mch:100, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> references:
#> row:13, col:1, text:NULL, mch:Inf, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> footer_left:
#> row:14, col:1, text:NULL, mch:Inf, x:0, y:0.5, hjust:0, vjust:0.5, rot:0,
#> footer_right:
#> row:14, col:3, text:NULL, mch:Inf, x:1, y:0.5, hjust:1, vjust:0.5, rot:0,
#> watermark:
#> row:10, col:2, text:NULL, mch:Inf, x:0.5, y:0.5, hjust:0.5, vjust:0.5, rot:45,
#> gpar - fontsize:80, alpha:0.3,
#>
library(magrittr)
# Customize margins and global graphical parameters and fill all cells
gridify(
object = ggplot2::ggplot(data = mtcars, ggplot2::aes(x = mpg, y = wt)) +
ggplot2::geom_line(),
layout = pharma_layout_base(
margin = grid::unit(c(0.5, 0.5, 0.5, 0.5), "inches"),
global_gpar = list(col = "blue", fontsize = 10)
)
) %>%
set_cell("header_left_1", "My Company") %>%
set_cell("header_left_2", "<PROJECT> / <INDICATION>") %>%
set_cell("header_left_3", "<STUDY>") %>%
set_cell("header_right_1", "CONFIDENTIAL") %>%
set_cell("header_right_2", "<Draft or Final>") %>%
set_cell("header_right_3", "Data Cut-off: YYYY-MM-DD") %>%
set_cell("output_num", "<Output> xx.xx.xx") %>%
set_cell("title_1", "<Title 1>") %>%
set_cell("title_2", "<Title 2>") %>%
set_cell("title_3", "<Optional Title 3>") %>%
set_cell("by_line", "By: <GROUP>, <optionally: Demographic parameters>") %>%
set_cell("note", "<Note or Footnotes>") %>%
set_cell("references", "<References:>") %>%
set_cell("footer_left", "Program: <PROGRAM NAME>, YYYY-MM-DD at HH:MM") %>%
set_cell("footer_right", "Page xx of nn") %>%
set_cell("watermark", "DRAFT")
#> gridifyClass object
#> ---------------------
#> Please run `show_spec(object)` or print the layout to get more specs.
#>
#> Cells:
#> header_left_1: filled
#> header_left_2: filled
#> header_left_3: filled
#> header_right_1: filled
#> header_right_2: filled
#> header_right_3: filled
#> output_num: filled
#> title_1: filled
#> title_2: filled
#> title_3: filled
#> by_line: filled
#> note: filled
#> references: filled
#> footer_left: filled
#> footer_right: filled
#> watermark: filled