library(admiral)
library(dplyr, warn.conflicts = FALSE)
library(tibble)
library(tidyverse)
library(lubridate){admiral 1.1.1} is out on CRAN! Though it may seem like just yesterday that {admiral} achieved its milestone 1.0 release, in actual fact six long months have gone by - testament to our commitment to now release twice-yearly rather than quarterly. During this time, we in the admiral team received lots of positive feedback from our community, but have nevertheless worked tirelessly to improve our package. This new release comes with various quality of life changes, targeted additions to our functions’ capabilities and improvements to our documentation that all contribute to improve each and every user’s experience - all while ensuring that our commitment to stability and a low amount of breaking changes is still met.
This blog post will showcase some of the highlights of this release, but you can explore the full contents in our Changelog.
Setup
The examples in this post require the following packages:
Improved error messaging
Our marquee update for {admiral} 1.1 is certainly our new and improved error messaging. Through a deep rework of our {admiraldev} package we have completely reworked how errors and warnings are thrown by {admiral} - all without creating any breaking changes for our users. In our new error messages:
- The name of the calling function appears on the top line to orient the user where the error is coming from.
- Color coding of words is now present so that the messages are easier on the eyes and easier to read.
- When applicable, the message is followed by an informative message (with the standard
iprefix) providing details on what to do to address the error. - Informative messages may include a hyperlink (🤯) of the code to run to get more details. Clicking the link will auto-run the code in your console and print the examples of incompatible inputs.
Try it out yourself by running the code below!
adsl <- tribble(
~USUBJID, ~TRTSDT, ~EOSDT,
"01", ymd("2020-12-06"), ymd("2021-03-06"),
"02", ymd("2021-01-16"), ymd("2021-02-03")
) %>%
mutate(STUDYID = "AB42")
ae <- tribble(
~USUBJID, ~AESTDTC, ~AESEQ, ~AEDECOD,
"01", "2021-01-03T10:56", 1, "Flu",
"01", "2021-03-04", 2, "Cough",
"01", "2021", 3, "Flu"
) %>%
mutate(STUDYID = "AB42")
ae_ext <- derive_vars_dt(
ae,
dtc = AESTDTC,
new_vars_prefix = "AEST",
highest_imputation = "M",
flag_imputation = "none"
)
ttae <- event_source(
dataset_name = "ae",
date = AESTDT,
set_values_to = exprs(
EVNTDESC = "AE",
SRCDOM = "AE",
SRCVAR = "AESTDTC",
SRCSEQ = AESEQ
)
)
eos <- censor_source(
dataset_name = "adsl",
date = EOSDT,
set_values_to = exprs(
EVNTDESC = "END OF STUDY",
SRCDOM = "ADSL",
SRCVAR = "EOSDT"
)
)
derive_param_tte(
dataset_adsl = adsl,
by_vars = exprs(AEDECOD),
start_date = TRTSDT,
event_conditions = list(ttae),
censor_conditions = list(eos),
source_datasets = list(adsl = adsl, ae = ae_ext),
set_values_to = exprs(
PARAMCD = "TTAE", # This is incorrect syntax! Not unique!
PARAM = paste("Time to First", AEDECOD, "Adverse Event"),
PARCAT1 = "TTAE",
PARCAT2 = AEDECOD
)
)
While we hope you don’t encounter too many errors and warnings while working with {admiral}, we also hope that when you do, our improved messaging will put you on the right path towards correcting them 😀.
New metadata object for ISO 3166 country codes
{admiral} now offers a new metadata object, country_code_lookup(), that can aid metadata added to decode countries based on ISO 3166 codes. Our hugely-flexible derive_vars_merged() then comes in handy if you ever need to leverage this metadata:
adsl <- tribble(
~USUBJID, ~COUNTRY,
"ST01-01", "AUT",
"ST01-02", "MWI",
"ST01-03", "GBR",
"ST01-04", "CHE",
"ST01-05", "NOR",
"ST01-06", "JPN",
"ST01-07", "USA"
)
adsl %>%
derive_vars_merged(
dataset_add = country_code_lookup,
new_vars = exprs(COUNTRYN = country_number, COUNTRYL = country_name),
by_vars = exprs(COUNTRY = country_code)
)# A tibble: 7 × 4
USUBJID COUNTRY COUNTRYN COUNTRYL
<chr> <chr> <dbl> <chr>
1 ST01-01 AUT 16 Austria
2 ST01-02 MWI 157 Malawi
3 ST01-03 GBR 80 United Kingdom of Great Britain and Northern Ireland
4 ST01-04 CHE 42 Switzerland
5 ST01-05 NOR 168 Norway
6 ST01-06 JPN 116 Japan
7 ST01-07 USA 235 United States of America If you were worried about having to type these up manually yourself, fear not as we now have you covered!
New helper functions for metadata-driven programming
Driven by a request from our GSK users, the {admiral} back-end now contains helper functions to derive_vars_merged() (get_flagged_records()) and derive_vars_query() (get_vars_query()) so that those can be called independently if required. This should aid efforts to automate execution of {admiral} code in a metadata-drive way. You can read more about the the reasoning behind this update in the following discussion on GitHub.
Enhancements to derive_var_trtemfl()
The function derive_var_trtemfl() now comes with a new (optional) argument, group_var. This allows users to derive Treatment Emergent Flags in Adverse Event ADaMs in the case where data are collected as one episode of AE with multiple lines. Indeed, if the argument is specified, derive_var_trtemfl() then assumes that AEs are recorded as one episode of AE with multiple lines using a grouping variable. Events starting during treatment or before treatment and worsening afterward are then flagged. Once an AE record in a group is flagged, all subsequent records in the treatment window are flagged regardless of severity. Take a look at the example below, which uses the grouping variable AEGRPID. In particular, observe how the second of the four AEs with AEGRPID = 2 occurs while on treatment and marks a worsening of severity since AETOXGR increases by one. As such, that AE and the following one are flagged as treatment emergent.
adae <- tribble(
~USUBJID, ~ASTDTM, ~AENDTM, ~AETOXGR, ~AEGRPID,
# before treatment
"1", ymd_hm("2021-12-13T20:15"), ymd_hm("2021-12-15T12:45"), "1", "1",
"1", ymd_hm("2021-12-14T20:15"), ymd_hm("2021-12-14T22:00"), "3", "1",
# starting before treatment and ending during treatment
"1", ymd_hm("2021-12-30T20:15"), ymd_hm("2022-01-14T01:23"), "3", "2",
"1", ymd_hm("2022-01-05T20:00"), ymd_hm("2022-06-01T11:00"), "1", "2",
"1", ymd_hm("2022-01-10T20:15"), ymd_hm("2022-01-11T01:23"), "2", "2",
"1", ymd_hm("2022-01-13T20:15"), ymd_hm("2022-03-01T01:23"), "1", "2",
# starting during treatment
"1", ymd_hm("2022-01-01T12:00"), ymd_hm("2022-01-02T23:25"), "4", "3"
) %>%
mutate(
TRTSDTM = ymd_hm("2022-01-01T01:01"),
TRTEDTM = ymd_hm("2022-04-30T23:59")
)
derive_var_trtemfl(
adae,
new_var = TRTEMFL,
trt_end_date = TRTEDTM,
end_window = 10,
intensity = AETOXGR,
group_var = AEGRPID,
subject_keys = exprs(USUBJID)
) %>%
select(ASTDTM, AENDTM, AETOXGR, AEGRPID, TRTEMFL)# A tibble: 7 × 5
ASTDTM AENDTM AETOXGR AEGRPID TRTEMFL
<dttm> <dttm> <chr> <chr> <chr>
1 2021-12-13 20:15:00 2021-12-15 12:45:00 1 1 <NA>
2 2021-12-14 20:15:00 2021-12-14 22:00:00 3 1 <NA>
3 2021-12-30 20:15:00 2022-01-14 01:23:00 3 2 <NA>
4 2022-01-05 20:00:00 2022-06-01 11:00:00 1 2 <NA>
5 2022-01-10 20:15:00 2022-01-11 01:23:00 2 2 Y
6 2022-01-13 20:15:00 2022-03-01 01:23:00 1 2 Y
7 2022-01-01 12:00:00 2022-01-02 23:25:00 4 3 Y Improved Visits and Periods vignette
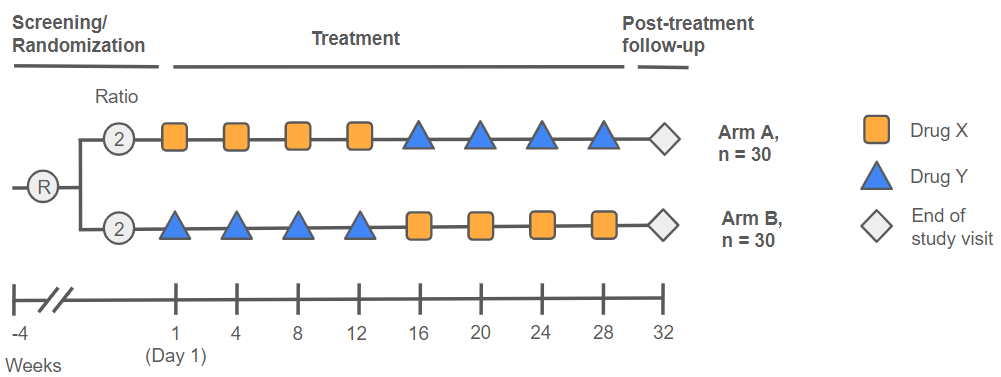
The Visits and Periods User Guide is one of {admiral}’s most popular - so much so that the authors of this post may or may not themselves refer to it often in their study work. The process of dealing with study periods and phases in particular can be complex and varies from study to study depending on design. Thus, {admiral} necessarily leaves some programming to be done ad-hoc by the ADaM programmer, but now with {admiral} 1.1 this vignette has been enhanced to include a worked example (complete with a dummy crossover design - see image below) to showcase how one may go about this difficult task. The vignette has also been reorganized for improved coherence.
New Get Started vignette
In this release cycle, we took the opportunity to review and revamp our Get Started vignette. The new and improved version of this page now places more focus on showcasing the main idea of {admiral}, i.e. constructing ADaM dataset by chaining together a series of modular blocks. Then, the page goes through some examples of the different types of blocks (that is, function calls) one can encounter, including adding variables to a dataset, adding records to a dataset and changing the nature of the existing datasets in some way.
New Programming Concepts and Conventions vignette
Coupled with the rework to the Get Started vignette, we now have a brand-new Programming Concepts and Conventions User Guide. Here, we aim to discuss some of the common programming concepts and conventions that have been adopted within the {admiral} family of packages, including how we handle missing values and how we work with expressions, quoting and unquoting. We hope this new page can assume encyclopaedic qualities for our users, especially for those topics like expressions that have an associated learning curve!
User Guide Reorganization
We took the opportunity to reorganize how our vignettes are categorized on the website, in order to present them in a more intuitive and tidy arrangement. They are now split into General Guides, ADaM Script Guides and Special Topics Guides. You can now see the full new arrangement below:
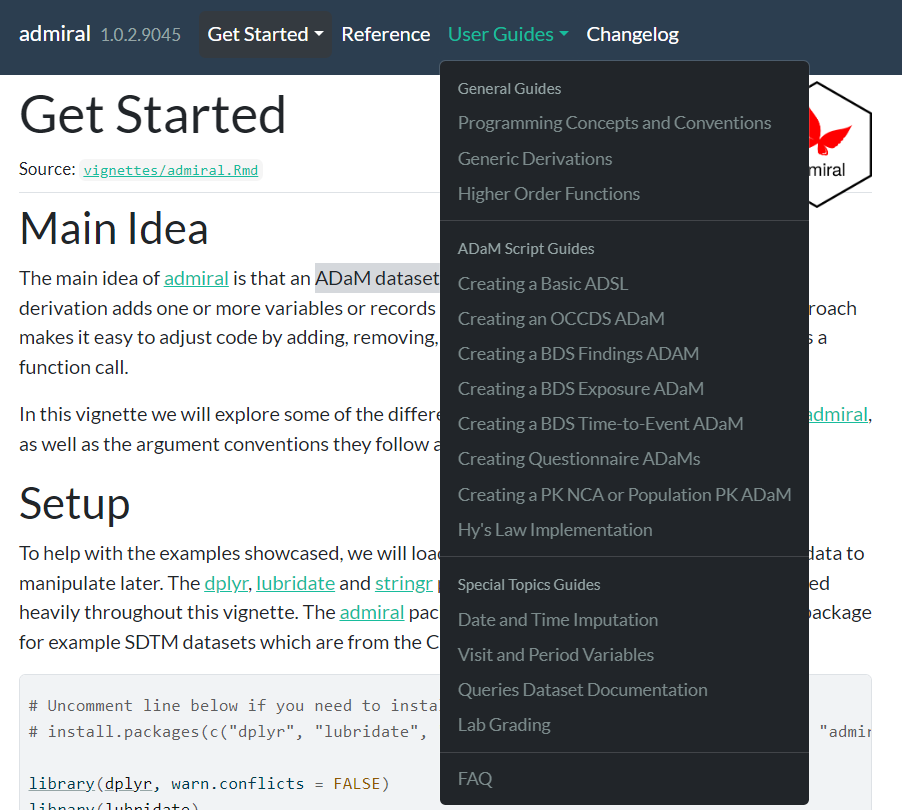
Let us know what you think of the new vignettes and organization of the website!
What’s next for admiral and the admiral family?
So, what’s next for {admiral} and the {admiral} family? For the {admiral} core package, starting in July we will be working towards an {admiral} 1.2 release in another six months time, but we want to hear from you, the {admiral} community! What do you think we should focus our efforts on, and what should be our priorities? let us know in the pharmaverse Slack, or by creating an issue or discussion in our GitHub repo.
On the package extension side, we have:
- New release of {admiralonco} (1.1) featuring a new vignette for creating ADRS with IMWG criteria (full changelog here).
- New release of {admiralophtha} (1.1) featuring improved ADOE, ADBCVA and ADVFQ templates (full changelog here).
- Upcoming release of {admiralvaccine} - further info TBC but you can take a look at our progress on our GitHub repo.
- Upcoming first release of {admiralpeds} in July, focusing mainly on child growth/development charts.
- Team kickoff for {admiralmetabolic}, where we aim to provide tools for the cardiovascular and metabolism therapeutic area. Our first release is scheduled in approximately six months time.
As you can see, progress in the {admiral} family never stops. We hope you’ve enjoyed this post, and that it’s inspired you to try out some of the new functionality. Happy ADaM programming!
Last updated
2026-01-23 20:54:18.713897
Details
Reuse
Citation
@online{mancini2024,
author = {Mancini, Edoardo and Straub, Ben},
title = {\{Admiral\} 1.1.1 Is Here!},
date = {2024-06-20},
url = {https://pharmaverse.github.io/blog/posts/2024-06-07_admiral_1.1_is.../admiral_1.1_is_here.html},
langid = {en}
}