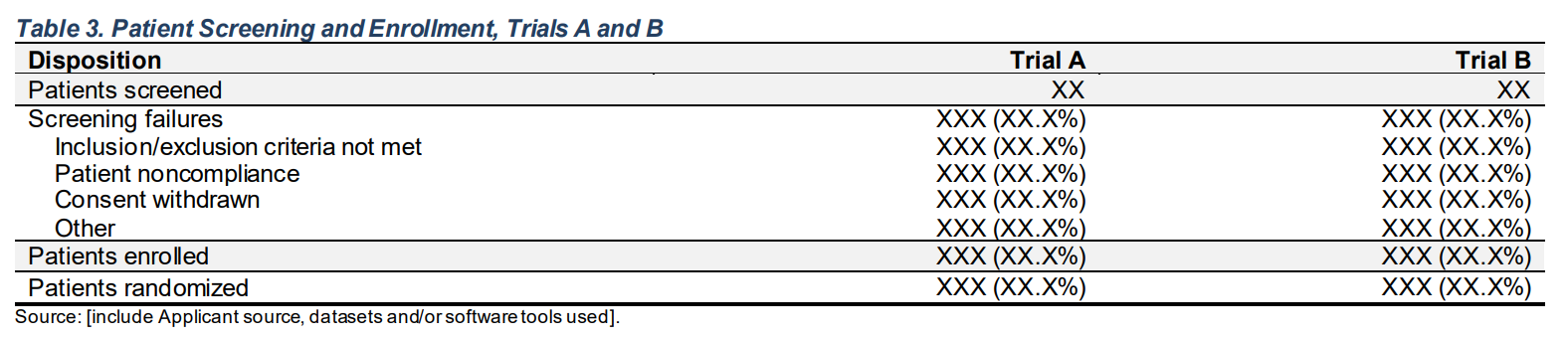
Disposition A: Drug X B: Placebo C: Combination
——————————————————————————————————————————————————————————————————————————————————
Patients screened 134 134 132
Screening failures 42 (31.3%) 32 (23.9%) 26 (19.7%)
Inclusion/exclusion criteria not met 6 (4.5%) 10 (7.5%) 10 (7.6%)
Patient noncompliance 12 (9.0%) 2 (1.5%) 6 (4.5%)
Consent withdrawn 13 (9.7%) 12 (9.0%) 5 (3.8%)
Other 11 (8.2%) 8 (6.0%) 5 (3.8%)
Patients enrolled 92 (68.7%) 102 (76.1%) 106 (80.3%)
Patients randomized 92 (68.7%) 102 (76.1%) 106 (80.3%) FDA Table 3
Patient Screening and Enrollment, Trials A and B
# Load Libraries & Data
library(dplyr)
library(cardinal)
adsl <- random.cdisc.data::cadsl
# Pre-Processing - Add all required screening variables are in df
set.seed(1)
adsl <- random.cdisc.data::cadsl
adsl$RANDDT[sample(seq_len(nrow(adsl)), 100)] <- NA
adsl <- adsl %>%
mutate(
ENRLDT = RANDDT,
SCRNFL = "Y",
SCRNFRS = factor(sample(
c("Inclusion/exclusion criteria not met", "Patient noncompliance", "Consent withdrawn", "Other"),
size = nrow(adsl), replace = TRUE
), levels = c("Inclusion/exclusion criteria not met", "Patient noncompliance", "Consent withdrawn", "Other")),
SCRNFAILFL = ifelse(is.na(ENRLDT), "Y", "N")
)
adsl$SCRNFRS[adsl$SCRNFL == "N" | !is.na(adsl$ENRLDT)] <- NA
# Output Table
make_table_03(df = adsl, scrnfl_var = "SCRNFL", scrnfailfl_var = "SCRNFAILFL", scrnfail_var = "SCRNFRS")make_table_3()
Required variables:
-
adsl:USUBJID,ENRLDT,RANDDT, and the variables specified byarm_var,scrnfl_var,scrnfailfl_var, andscrnfail_var. -
alt_counts_df(if specified):USUBJID, and the variable specified byarm_var.
| Argument | Description | Default |
|---|---|---|
adsl |
(data.frame) Dataset (typically ADSL) required to build table. |
No default |
alt_counts_df |
(character) Alternative dataset used only to calculate column counts. |
NULL |
show_colcounts |
(flag) Whether column counts should be printed. |
FALSE |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
scrnfl_var |
(character) Variable from df that indicates whether patients were screened. |
No default |
scrnfailfl_var |
(character) Variable from df that indicates screening failure. |
No default |
scrnfail_var |
(character) Variable from df that contains reasons for screening failure. |
No default |
lbl_overall |
(character) If specified, an overall column will be added to the table with the given value as the column label. |
NULL |
prune_0 |
(flag) Whether all-zero rows should be removed from the table. |
TRUE |
annotations |
(named list of character) List of annotations to add to the table. Valid annotation types are title, subtitles, main_footer, and prov_footer. Each name-value pair should use the annotation type as name and the desired string as value. |
NULL |
Source code for this function is available here.