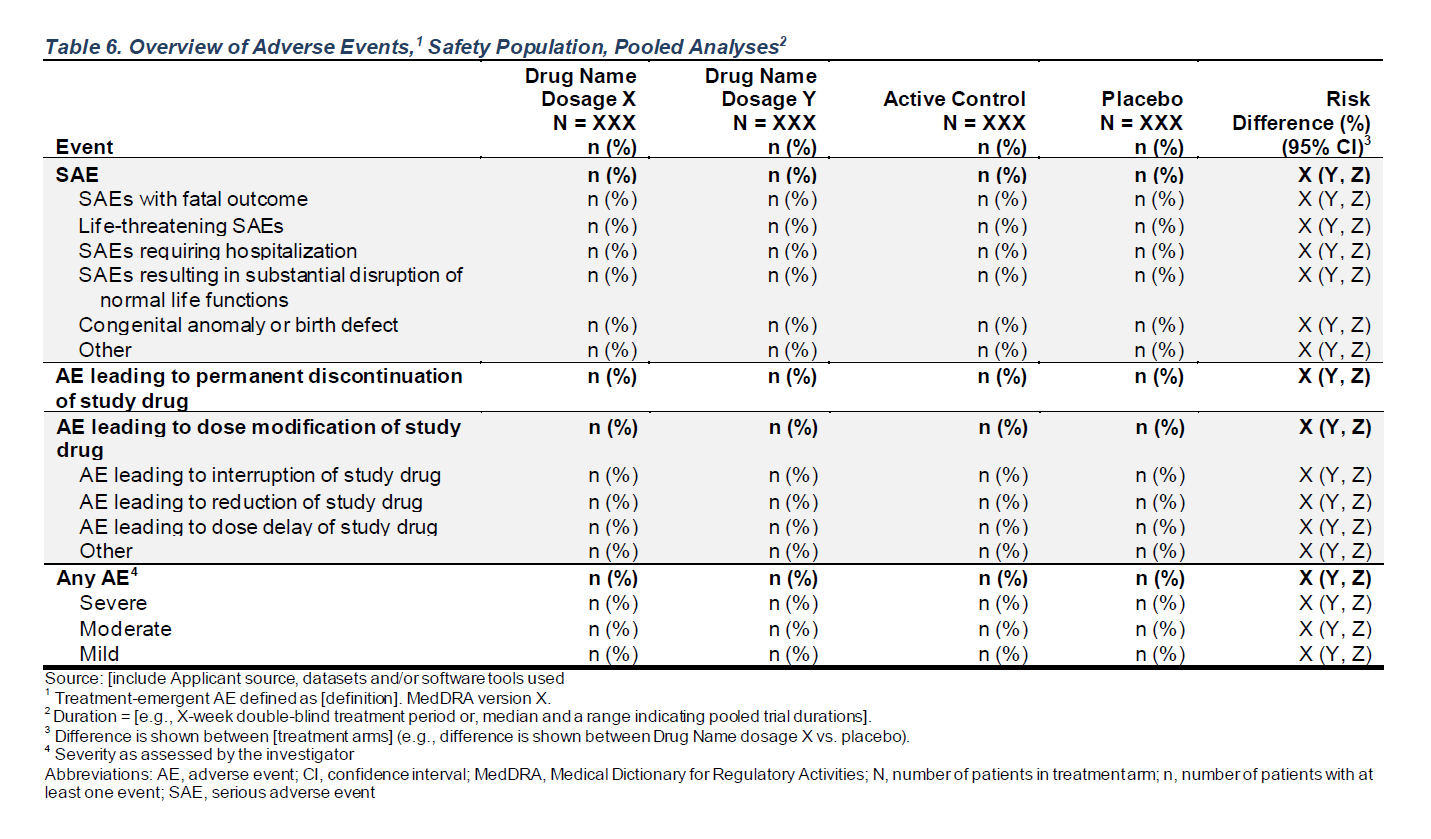
FDA Table 6
Overview of Adverse Events, Safety Population, Pooled Analyses
gtsummary Table Setup
| Event |
A: Drug X N = 1341 |
B: Placebo N = 1341 |
C: Combination N = 1321 |
|---|---|---|---|
| SAE | 104 (78%) | 101 (75%) | 99 (75%) |
| SAEs with fatal outcome | 76 (57%) | 70 (52%) | 75 (57%) |
| Life-threatening SAEs | 9 (6.7%) | 13 (9.7%) | 19 (14%) |
| SAEs requiring hospitalization | 24 (18%) | 28 (21%) | 30 (23%) |
| SAEs resulting in substantial disruption of normal life functions | 28 (21%) | 21 (16%) | 23 (17%) |
| Congenital anomaly or birth defect | 26 (19%) | 27 (20%) | 20 (15%) |
| Other | 30 (22%) | 37 (28%) | 32 (24%) |
| AE leading to permanent discontinuation of study drug | 75 (56%) | 73 (54%) | 83 (63%) |
| AE leading to dose modification of study drug | 71 (53%) | 86 (64%) | 83 (63%) |
| AE leading to dose delay of study drug | 20 (15%) | 28 (21%) | 30 (23%) |
| AE leading to interruption of study drug | 4 (3.0%) | 4 (3.0%) | 3 (2.3%) |
| AE leading to reduction of study drug | 46 (34%) | 46 (34%) | 51 (39%) |
| Other | 34 (25%) | 48 (36%) | 46 (35%) |
| Any AE | 122 (91%) | 123 (92%) | 120 (91%) |
| MILD | 88 (66%) | 95 (71%) | 102 (77%) |
| MODERATE | 99 (74%) | 100 (75%) | 103 (78%) |
| SEVERE | 91 (68%) | 90 (67%) | 93 (70%) |
| 1 n (%) | |||
Function Details
make_table_06()
Required variables:
-
df: The variables specified byid_varandarm_var. -
denominator: The variables specified bysaffl_var,arm_var,sae_var,sae_cat_vars,disc_var,dose_mod_var, andsev_var.
| Argument | Description | Default |
df |
(data.frame) Dataset (typically ADAE) required to build the table. |
No default |
return_ard |
(flag) Whether an ARD should be returned. |
TRUE |
denominator |
(character) Alternative dataset (typically ADSL) used only to calculate column counts. |
No default |
id_var |
(character) Identifier variable used to count the participants within each category. |
"USUBJID" |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
sae_var |
(character) Flag variable used to indicate whether an adverse event is serious or not. |
"AESER" |
sae_cat_vars |
(character list) Named list of flag variables for categories of serious adverse events. List names represent the labels in the table. |
"list("SAEs with fatal outcome" = "AESDTH", "Life-threatening SAEs" = "AESLIFE", "SAEs requiring hospitalization" = "AESHOSP", "SAEs resulting in substantial disruption of normal life functions" = "AESDISAB", "Congenital anomaly or birth defect" = "AESCONG", "Other" = "AESMIE")" |
disc_var |
(character) Flag variable used to indicate whether the subject was discontinued from the study as result of an adverse event. |
"AEACNOTH" |
dose_mod_var |
(character) Flag variable used to indicate whether the dose was modified as result of an adverse event. |
"AEACN" |
dose_mod_cat_labels |
(character list) Named list of labels. Names may be “DRUG INTERRUPTED”, “DOSE REDUCED”, “DOSE RATE REDUCED”, “DOSE INCREASED”. |
"list("DRUG INTERRUPTED" = "AE leading to interruption of study drug", "DOSE REDUCED" = "AE leading to reduction of study drug" , "DOSE RATE REDUCED" = "AE leading to dose delay of study drug", "DOSE INCREASED" = "Other")" |
sev_var |
(character) Variable containing the severity of an adverse event. |
"AESEV" |
na_level |
(character) String to represent missing values. |
"<Missing>" |
ARD Setup
[[1]]
[[1]][[1]]
[[1]][[1]]$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESER SAE hierarch… n n 104 104 <fn> stat_1
11 ARM A: Drug X AESER SAE hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESER SAE hierarch… p % 0.776 78 <fn> stat_1
13 ARM B: Place… AESER SAE hierarch… n n 101 101 <fn> stat_2
14 ARM B: Place… AESER SAE hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESER SAE hierarch… p % 0.754 75 <fn> stat_2
16 ARM C: Combi… AESER SAE hierarch… n n 99 99 <fn> stat_3
17 ARM C: Combi… AESER SAE hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESER SAE hierarch… p % 0.75 75 <fn> stat_3
[[1]]$`SAEs with fatal outcome`
[[1]]$`SAEs with fatal outcome`$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESDTH SAEs wit… hierarch… n n 76 76 <fn> stat_1
11 ARM A: Drug X AESDTH SAEs wit… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESDTH SAEs wit… hierarch… p % 0.567 57 <fn> stat_1
13 ARM B: Place… AESDTH SAEs wit… hierarch… n n 70 70 <fn> stat_2
14 ARM B: Place… AESDTH SAEs wit… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESDTH SAEs wit… hierarch… p % 0.522 52 <fn> stat_2
16 ARM C: Combi… AESDTH SAEs wit… hierarch… n n 75 75 <fn> stat_3
17 ARM C: Combi… AESDTH SAEs wit… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESDTH SAEs wit… hierarch… p % 0.568 57 <fn> stat_3
[[1]]$`Life-threatening SAEs`
[[1]]$`Life-threatening SAEs`$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESLIFE Life-thr… hierarch… n n 9 9 <fn> stat_1
11 ARM A: Drug X AESLIFE Life-thr… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESLIFE Life-thr… hierarch… p % 0.067 6.7 <fn> stat_1
13 ARM B: Place… AESLIFE Life-thr… hierarch… n n 13 13 <fn> stat_2
14 ARM B: Place… AESLIFE Life-thr… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESLIFE Life-thr… hierarch… p % 0.097 9.7 <fn> stat_2
16 ARM C: Combi… AESLIFE Life-thr… hierarch… n n 19 19 <fn> stat_3
17 ARM C: Combi… AESLIFE Life-thr… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESLIFE Life-thr… hierarch… p % 0.144 14 <fn> stat_3
[[1]]$`SAEs requiring hospitalization`
[[1]]$`SAEs requiring hospitalization`$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESHOSP SAEs req… hierarch… n n 24 24 <fn> stat_1
11 ARM A: Drug X AESHOSP SAEs req… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESHOSP SAEs req… hierarch… p % 0.179 18 <fn> stat_1
13 ARM B: Place… AESHOSP SAEs req… hierarch… n n 28 28 <fn> stat_2
14 ARM B: Place… AESHOSP SAEs req… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESHOSP SAEs req… hierarch… p % 0.209 21 <fn> stat_2
16 ARM C: Combi… AESHOSP SAEs req… hierarch… n n 30 30 <fn> stat_3
17 ARM C: Combi… AESHOSP SAEs req… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESHOSP SAEs req… hierarch… p % 0.227 23 <fn> stat_3
[[1]]$`SAEs resulting in substantial disruption of normal life functions`
[[1]]$`SAEs resulting in substantial disruption of normal life functions`$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESDISAB SAEs res… hierarch… n n 28 28 <fn> stat_1
11 ARM A: Drug X AESDISAB SAEs res… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESDISAB SAEs res… hierarch… p % 0.209 21 <fn> stat_1
13 ARM B: Place… AESDISAB SAEs res… hierarch… n n 21 21 <fn> stat_2
14 ARM B: Place… AESDISAB SAEs res… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESDISAB SAEs res… hierarch… p % 0.157 16 <fn> stat_2
16 ARM C: Combi… AESDISAB SAEs res… hierarch… n n 23 23 <fn> stat_3
17 ARM C: Combi… AESDISAB SAEs res… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESDISAB SAEs res… hierarch… p % 0.174 17 <fn> stat_3
[[1]]$`Congenital anomaly or birth defect`
[[1]]$`Congenital anomaly or birth defect`$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESCONG Congenit… hierarch… n n 26 26 <fn> stat_1
11 ARM A: Drug X AESCONG Congenit… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESCONG Congenit… hierarch… p % 0.194 19 <fn> stat_1
13 ARM B: Place… AESCONG Congenit… hierarch… n n 27 27 <fn> stat_2
14 ARM B: Place… AESCONG Congenit… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESCONG Congenit… hierarch… p % 0.201 20 <fn> stat_2
16 ARM C: Combi… AESCONG Congenit… hierarch… n n 20 20 <fn> stat_3
17 ARM C: Combi… AESCONG Congenit… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESCONG Congenit… hierarch… p % 0.152 15 <fn> stat_3
[[1]]$Other
[[1]]$Other$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AESMIE Other hierarch… n n 30 30 <fn> stat_1
11 ARM A: Drug X AESMIE Other hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AESMIE Other hierarch… p % 0.224 22 <fn> stat_1
13 ARM B: Place… AESMIE Other hierarch… n n 37 37 <fn> stat_2
14 ARM B: Place… AESMIE Other hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AESMIE Other hierarch… p % 0.276 28 <fn> stat_2
16 ARM C: Combi… AESMIE Other hierarch… n n 32 32 <fn> stat_3
17 ARM C: Combi… AESMIE Other hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AESMIE Other hierarch… p % 0.242 24 <fn> stat_3
[[2]]
[[2]]$tbl_hierarchical{cards} data frame: 18 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X AEACNOTH AE leadi… hierarch… n n 75 75 <fn> stat_1
11 ARM A: Drug X AEACNOTH AE leadi… hierarch… N N 134 134 <fn> stat_1
12 ARM A: Drug X AEACNOTH AE leadi… hierarch… p % 0.56 56 <fn> stat_1
13 ARM B: Place… AEACNOTH AE leadi… hierarch… n n 73 73 <fn> stat_2
14 ARM B: Place… AEACNOTH AE leadi… hierarch… N N 134 134 <fn> stat_2
15 ARM B: Place… AEACNOTH AE leadi… hierarch… p % 0.545 54 <fn> stat_2
16 ARM C: Combi… AEACNOTH AE leadi… hierarch… n n 83 83 <fn> stat_3
17 ARM C: Combi… AEACNOTH AE leadi… hierarch… N N 132 132 <fn> stat_3
18 ARM C: Combi… AEACNOTH AE leadi… hierarch… p % 0.629 63 <fn> stat_3
[[3]]
[[3]]$tbl_hierarchical{cards} data frame: 54 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X ..ard_hierarchical_overall.. TRUE hierarch… n n 71 71 <fn> stat_1ℹ 44 more rowsℹ Use `print(n = ...)` to see more rows
[[4]]
[[4]]$tbl_hierarchical{cards} data frame: 45 x 13 group1 group1_level variable variable_level context stat_name stat_label stat stat_fmt fmt_fn warning error gts_column
1 <NA> ARM A: Drug X categori… n n 134 134 0 stat_1
2 <NA> ARM A: Drug X categori… N N 400 400 0 stat_1
3 <NA> ARM A: Drug X categori… p % 0.335 33.5 <fn> stat_1
4 <NA> ARM B: Place… categori… n n 134 134 0 stat_2
5 <NA> ARM B: Place… categori… N N 400 400 0 stat_2
6 <NA> ARM B: Place… categori… p % 0.335 33.5 <fn> stat_2
7 <NA> ARM C: Combi… categori… n n 132 132 0 stat_3
8 <NA> ARM C: Combi… categori… N N 400 400 0 stat_3
9 <NA> ARM C: Combi… categori… p % 0.33 33.0 <fn> stat_3
10 ARM A: Drug X ..ard_hierarchical_overall.. TRUE hierarch… n n 122 122 <fn> stat_1ℹ 35 more rows
ℹ Use `print(n = ...)` to see more rowsrtables Table Setup
A: Drug X B: Placebo C: Combination Risk Difference (%) (95% CI)
Event (N=134) (N=134) (N=132) (N=268)
———————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
SAE 104 (85.2%) 101 (82.1%) 99 (82.5%) -2.2 (-12.4 - 7.9)
SAEs with fatal outcome 76 (62.3%) 70 (56.9%) 75 (62.5%) -4.5 (-16.4 - 7.4)
Life-threatening SAEs 9 (7.4%) 13 (10.6%) 19 (15.8%) 3.0 (-3.6 - 9.5)
SAEs requiring hospitalization 24 (19.7%) 28 (22.8%) 30 (25.0%) 3.0 (-6.5 - 12.4)
SAEs resulting in substantial disruption of normal life functions 28 (23.0%) 21 (17.1%) 23 (19.2%) -5.2 (-14.5 - 4.0)
Congenital anomaly or birth defect 26 (21.3%) 27 (22.0%) 20 (16.7%) 0.7 (-8.8 - 10.3)
Other 30 (24.6%) 37 (30.1%) 32 (26.7%) 5.2 (-5.1 - 15.6)
AE leading to permanent discontinuation of study drug 27 (22.1%) 26 (21.1%) 30 (25.0%) -0.7 (-10.3 - 8.8)
AE leading to dose modification of study drug 71 (58.2%) 86 (69.9%) 83 (69.2%) 11.2 (-0.5 - 22.9)
AE leading to interruption of study drug 4 (3.3%) 4 (3.3%) 3 (2.5%) 0.0 (-4.1 - 4.1)
AE leading to reduction of study drug 46 (37.7%) 46 (37.4%) 51 (42.5%) 0.0 (-11.4 - 11.4)
AE leading to dose delay of study drug 20 (16.4%) 28 (22.8%) 30 (25.0%) 6.0 (-3.2 - 15.1)
Other 34 (27.9%) 48 (39.0%) 46 (38.3%) 10.4 (-0.5 - 21.4)
Any AE 122 (91.0%) 123 (91.8%) 120 (90.9%) 0.7 (-6.0 - 7.5)
MILD 7 (5.2%) 9 (6.7%) 4 (3.0%) 1.5 (-4.2 - 7.2)
MODERATE 24 (17.9%) 24 (17.9%) 23 (17.4%) 0.0 (-9.2 - 9.2)
SEVERE 91 (67.9%) 90 (67.2%) 93 (70.5%) -0.7 (-12.0 - 10.5) Function Details
make_table_06_rtables()
Required variables:
-
adae:USUBJID,TRTEMFL,AESEV,AESER,AESDTH,AESLIFE,AESHOSP,AESDISAB,AESCONG,AESMIE,AEACN, and the variables specified byarm_varandsaffl_var. -
alt_counts_df(if specified):USUBJIDand the variables specified byarm_varandsaffl_var.
| Argument | Description | Default |
adae |
(data.frame) Dataset (typically ADAE) required to build table. |
No default |
alt_counts_df |
(character) Alternative dataset (typically ADSL) used only to calculate column counts. |
NULL |
show_colcounts |
(flag) Whether column counts should be printed. |
TRUE |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
lbl_overall |
(character) If specified, an overall column will be added to the table with the given value as the column label. |
NULL |
risk_diff |
(named
|
NULL |
prune_0 |
(flag) Whether all-zero rows should be removed from the table. |
FALSE |
annotations |
(named list of character) List of annotations to add to the table. Valid annotation types are title, subtitles, main_footer, and prov_footer. Each name-value pair should use the annotation type as name and the desired string as value. |
NULL |