|
Body System or Organ Class FMQ (Narrow) |
A: Drug X N = 1341 |
B: Placebo N = 1341 |
C: Combination N = 1321 |
|---|---|---|---|
| cl A.1 | |||
| FMQ1 | 17 (13%) | 14 (10%) | 29 (22%) |
| FMQ2 | 23 (17%) | 20 (15%) | 20 (15%) |
| FMQ3 | 20 (15%) | 19 (14%) | 23 (17%) |
| cl B.1 | |||
| FMQ1 | 8 (6.0%) | 11 (8.2%) | 7 (5.3%) |
| FMQ2 | 5 (3.7%) | 12 (9.0%) | 16 (12%) |
| FMQ3 | 10 (7.5%) | 7 (5.2%) | 5 (3.8%) |
| cl B.2 | |||
| FMQ1 | 13 (9.7%) | 15 (11%) | 9 (6.8%) |
| FMQ2 | 12 (9.0%) | 9 (6.7%) | 10 (7.6%) |
| FMQ3 | 6 (4.5%) | 7 (5.2%) | 9 (6.8%) |
| cl C.2 | |||
| FMQ1 | 9 (6.7%) | 9 (6.7%) | 12 (9.1%) |
| FMQ2 | 6 (4.5%) | 8 (6.0%) | 8 (6.1%) |
| FMQ3 | 6 (4.5%) | 9 (6.7%) | 10 (7.6%) |
| cl D.1 | |||
| FMQ1 | 23 (17%) | 17 (13%) | 27 (20%) |
| FMQ2 | 22 (16%) | 20 (15%) | 25 (19%) |
| FMQ3 | 15 (11%) | 19 (14%) | 21 (16%) |
| cl D.2 | |||
| FMQ1 | 8 (6.0%) | 11 (8.2%) | 9 (6.8%) |
| FMQ2 | 14 (10%) | 15 (11%) | 14 (11%) |
| FMQ3 | 11 (8.2%) | 9 (6.7%) | 11 (8.3%) |
| 1 n (%) | |||
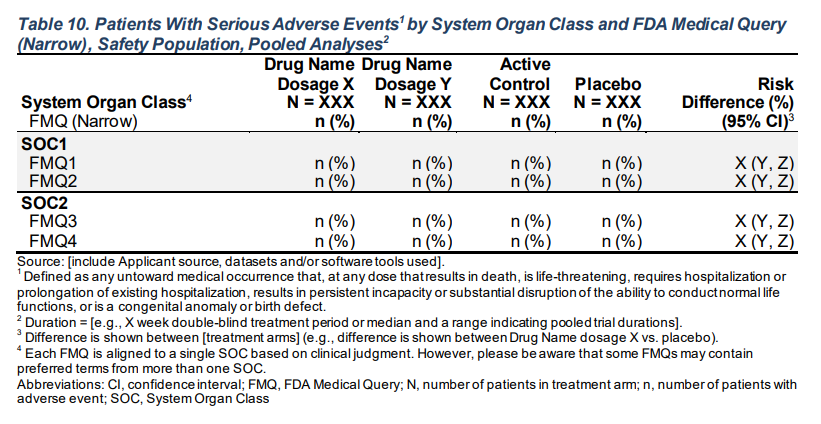
FDA Table 10
Patients With Serious Adverse Events by System Organ Class and FDA Medical Query (Narrow), Safety Population, Pooled Analyses
# Load Libraries & Data
library(dplyr)
library(cards)
library(cardinal)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Pre-Processing - Ensure required variables fmqsc_var and fmqnam_var exist in adae
set.seed(1)
adae <- adae %>%
rename(FMQ01SC = SMQ01SC) %>%
mutate(
AESER = sample(c("Y", "N"), size = nrow(adae), replace = TRUE),
FMQ01NAM = sample(c("FMQ1", "FMQ2", "FMQ3"), size = nrow(adae), replace = TRUE)
)
adae$FMQ01SC[is.na(adae$FMQ01SC)] <- "NARROW"
# Output Table
make_table_10(df = adae, denominator = adsl, return_ard = FALSE)make_table_10()
Required variables:
-
df:AEBODSYS,AESER, and the variables specified byid_var,arm_var,saffl_var,fmqsc_var, andfmqnam_var. -
denominator(if specified): The variables specified byid_var,arm_varandsaffl_var.
| Argument | Description | Default |
df |
(data.frame) Dataset (typically ADAE) required to build table. |
No default |
denominator |
(character) Alternative dataset used only to calculate column counts. |
NULL |
return_ard |
(flag) Whether column counts should be printed. |
TRUE |
id_var |
(character) Name of the unique subject identifiers variable. |
"USUBJID" |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
fmqsc_var |
(character) FMQ scope variable to use in table. |
"FMQ01SC" |
fmqnam_var |
(character) FMQ reference name variable to use in table. |
"FMQ01NAM" |
fmq_scope |
(character) FMQ scope, can be ‘“NARROW”’ or ‘“BROAD”’. |
"NARROW" |
na_level |
(character) String to represent missing values. |
"<Missing>" |
Source code for this function is available here.
| group1 | group1_level | group2 | group2_level | variable | variable_level | context | stat_name | stat_label | stat | fmt_fn | warning | error |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | n | n | 17 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | p | % | 0.1268657 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | n | n | 23 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | p | % | 0.1716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | n | n | 20 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | p | % | 0.1492537 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | n | n | 8 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | p | % | 0.05970149 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | n | n | 5 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | p | % | 0.03731343 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | n | n | 10 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | p | % | 0.07462687 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | n | n | 13 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | p | % | 0.09701493 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | n | n | 12 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | p | % | 0.08955224 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | n | n | 6 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | p | % | 0.04477612 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | p | % | 0.06716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | n | n | 6 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | p | % | 0.04477612 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | n | n | 6 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | p | % | 0.04477612 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | n | n | 23 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | p | % | 0.1716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | n | n | 22 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | p | % | 0.1641791 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | n | n | 15 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | p | % | 0.1119403 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | n | n | 8 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | p | % | 0.05970149 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | n | n | 14 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | p | % | 0.1044776 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | n | n | 11 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 1 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | p | % | 0.08208955 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | n | n | 14 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | p | % | 0.1044776 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | n | n | 20 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | p | % | 0.1492537 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | n | n | 19 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | p | % | 0.141791 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | n | n | 11 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | p | % | 0.08208955 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | n | n | 12 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | p | % | 0.08955224 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | n | n | 7 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | p | % | 0.05223881 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | n | n | 15 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | p | % | 0.1119403 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | p | % | 0.06716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | n | n | 7 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | p | % | 0.05223881 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | p | % | 0.06716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | n | n | 8 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | p | % | 0.05970149 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | p | % | 0.06716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | n | n | 17 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | p | % | 0.1268657 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | n | n | 20 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | p | % | 0.1492537 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | n | n | 19 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | p | % | 0.141791 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | n | n | 11 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | p | % | 0.08208955 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | n | n | 15 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | p | % | 0.1119403 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | N | N | 134 | 0 | NULL | NULL |
| ARM | 2 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | p | % | 0.06716418 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | n | n | 29 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 1 | hierarchical | p | % | 0.219697 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | n | n | 20 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 2 | hierarchical | p | % | 0.1515152 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | n | n | 23 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 1 | FMQ01NAM | 3 | hierarchical | p | % | 0.1742424 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | n | n | 7 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 1 | hierarchical | p | % | 0.0530303 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | n | n | 16 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 2 | hierarchical | p | % | 0.1212121 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | n | n | 5 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 2 | FMQ01NAM | 3 | hierarchical | p | % | 0.03787879 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 1 | hierarchical | p | % | 0.06818182 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | n | n | 10 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 2 | hierarchical | p | % | 0.07575758 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 3 | FMQ01NAM | 3 | hierarchical | p | % | 0.06818182 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | n | n | 12 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 1 | hierarchical | p | % | 0.09090909 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | n | n | 8 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 2 | hierarchical | p | % | 0.06060606 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | n | n | 10 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 5 | FMQ01NAM | 3 | hierarchical | p | % | 0.07575758 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | n | n | 27 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 1 | hierarchical | p | % | 0.2045455 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | n | n | 25 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 2 | hierarchical | p | % | 0.1893939 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | n | n | 21 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 6 | FMQ01NAM | 3 | hierarchical | p | % | 0.1590909 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | n | n | 9 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 1 | hierarchical | p | % | 0.06818182 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | n | n | 14 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 2 | hierarchical | p | % | 0.1060606 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | n | n | 11 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | N | N | 132 | 0 | NULL | NULL |
| ARM | 3 | AEBODSYS | 7 | FMQ01NAM | 3 | hierarchical | p | % | 0.08333333 | function (x) , {, res <- ifelse(is.na(x), NA_character_, str_trim(format(round_fun(x * , scale, digits = digits), nsmall = digits))), if (!is.null(width)) {, res <- ifelse(nchar(res) >= width | is.na(res), res, , paste0(strrep(” “, width - nchar(res)), res)), }, res, } | NULL | NULL |
# Load Libraries & Data
library(dplyr)
library(cards)
library(cardinal)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Pre-Processing - Ensure required variables fmqsc_var and fmqnam_var exist in adae
set.seed(1)
adae <- adae %>%
rename(FMQ01SC = SMQ01SC) %>%
mutate(
AESER = sample(c("Y", "N"), size = nrow(adae), replace = TRUE),
FMQ01NAM = sample(c("FMQ1", "FMQ2", "FMQ3"), size = nrow(adae), replace = TRUE)
)
adae$FMQ01SC[is.na(adae$FMQ01SC)] <- "NARROW"
# Output ARD
output <- make_table_10(df = adae, denominator = adsl)
output$ardBody System or Organ Class A: Drug X B: Placebo C: Combination
FMQ (Narrow) (N=134) (N=134) (N=132)
—————————————————————————————————————————————————————————————————————
cl A.1
FMQ1 17 (12.7%) 14 (10.4%) 29 (22.0%)
FMQ2 23 (17.2%) 20 (14.9%) 20 (15.2%)
FMQ3 20 (14.9%) 19 (14.2%) 23 (17.4%)
cl B.1
FMQ1 8 (6.0%) 11 (8.2%) 7 (5.3%)
FMQ2 5 (3.7%) 12 (9.0%) 16 (12.1%)
FMQ3 10 (7.5%) 7 (5.2%) 5 (3.8%)
cl B.2
FMQ1 13 (9.7%) 15 (11.2%) 9 (6.8%)
FMQ2 12 (9.0%) 9 (6.7%) 10 (7.6%)
FMQ3 6 (4.5%) 7 (5.2%) 9 (6.8%)
cl C.2
FMQ1 9 (6.7%) 9 (6.7%) 12 (9.1%)
FMQ2 6 (4.5%) 8 (6.0%) 8 (6.1%)
FMQ3 6 (4.5%) 9 (6.7%) 10 (7.6%)
cl D.1
FMQ1 23 (17.2%) 17 (12.7%) 27 (20.5%)
FMQ2 22 (16.4%) 20 (14.9%) 25 (18.9%)
FMQ3 15 (11.2%) 19 (14.2%) 21 (15.9%)
cl D.2
FMQ1 8 (6.0%) 11 (8.2%) 9 (6.8%)
FMQ2 14 (10.4%) 15 (11.2%) 14 (10.6%)
FMQ3 11 (8.2%) 9 (6.7%) 11 (8.3%) # Load Libraries & Data
# library(dplyr)
library(cardinal)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Pre-Processing - Ensure required variables fmqsc_var and fmqnam_var exist in adae
set.seed(1)
adae <- adae %>%
rename(FMQ01SC = SMQ01SC) %>%
mutate(
AESER = sample(c("Y", "N"), size = nrow(adae), replace = TRUE),
FMQ01NAM = sample(c("FMQ1", "FMQ2", "FMQ3"), size = nrow(adae), replace = TRUE)
)
adae$FMQ01SC[is.na(adae$FMQ01SC)] <- "NARROW"
# Output Table
make_table_10_rtables(df = adae, alt_counts_df = adsl)make_table_10_rtables()
Required variables:
-
df:AEBODSYS,AESER, and the variables specified byid_var,arm_var,saffl_var,fmqsc_var, andfmqnam_var. -
alt_counts_df(if specified): The variables specified byid_var,arm_varandsaffl_var.
| Argument | Description | Default |
df |
(data.frame) Dataset (typically ADAE) required to build table. |
No default |
alt_counts_df |
(character) Alternative dataset used only to calculate column counts. |
NULL |
show_colcounts |
(flag) Whether column counts should be printed. |
TRUE |
id_var |
(character) Name of the unique subject identifiers variable. |
"ARM" |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
fmqsc_var |
(character) FMQ scope variable to use in table. |
"FMQ01SC" |
fmqnam_var |
(character) FMQ reference name variable to use in table. |
"FMQ01NAM" |
fmq_scope |
(character) FMQ scope, can be ‘“NARROW”’ or ‘“BROAD”’. |
"NARROW" |
lbl_overall |
(character) If specified, an overall column will be added to the table with the given value as the column label. |
NULL |
risk_diff |
(named
|
NULL |
prune_0 |
(flag) Whether all-zero rows should be removed from the table. |
FALSE |
na_level |
(character) String to represent missing values. |
"<Missing>" |
annotations |
(named list of character) List of annotations to add to the table. Valid annotation types are title, subtitles, main_footer, and prov_footer. Each name-value pair should use the annotation type as name and the desired string as value. |
NULL |
Source code for this function is available here.