adae: adae must contain USUBJID, SEX, and the variables specified by arm_var, saffl_var, pref_var, fmqsc_var and fmqnam_var.
alt_counts_df (if specified): USUBJID and the variables specified by arm_var and saffl_var.
Argument
Description
Default
adae
(data.frame) Dataset (typically ADAE) required to build table.
No default
alt_counts_df
(character) Alternative dataset used only to calculate column counts.
NULL
show_colcounts
(flag) Whether column counts should be printed.
TRUE
arm_var
(character) Arm variable used to split table into columns.
"ARM"
saffl_var
(character) Flag variable used to indicate inclusion in safety population.
"SAFFL"
pref_var
(character) Preferred term variable to use in table.
"AEDECOD"
sex_scope
(character) Level of SEX to output in table.
"F"
fmq_scope
(character) FMQ scope value to use in table.
"BROAD"
fmqsc_var
(character) FMQ scope variable to use in table.
"FMQ01SC"
fmqnam_var
(character) FMQ reference name variable to use in table.
"FMQ01NAM"
lbl_overall
(character) If specified, an overall column will be added to the table with the given value as the column label.
NULL
risk_diff
(named list) List of settings to apply to add a risk difference column to the table. See tern::add_riskdiff() for more details. List should contain the following elements:
arm_x: (required) the name of reference arm.
arm_y: (required) the name of the arm to compare to the reference arm.
col_label: (optional) label to use for the risk difference column. Defaults to "Risk Difference (%) (95% CI)".
pct: (optional) whether the output should be returned as percentages. Defaults to TRUE.
NULL
prune_0
(flag) Whether all-zero rows should be removed from the table.
FALSE
na_level
(character) String to represent missing values.
"<Missing>"
annotations
(named list of character) List of annotations to add to the table. Valid annotation types are title, subtitles, main_footer, and prov_footer. Each name-value pair should use the annotation type as name and the desired string as value.
---title: FDA Table 18subtitle: Patients With Adverse Events by Female-Specific FDA Medical Query (Broad) and Preferred Term, Female Safety Population, Pooled Analysesformat: html---::: panel-tabset## Spec. Screenshot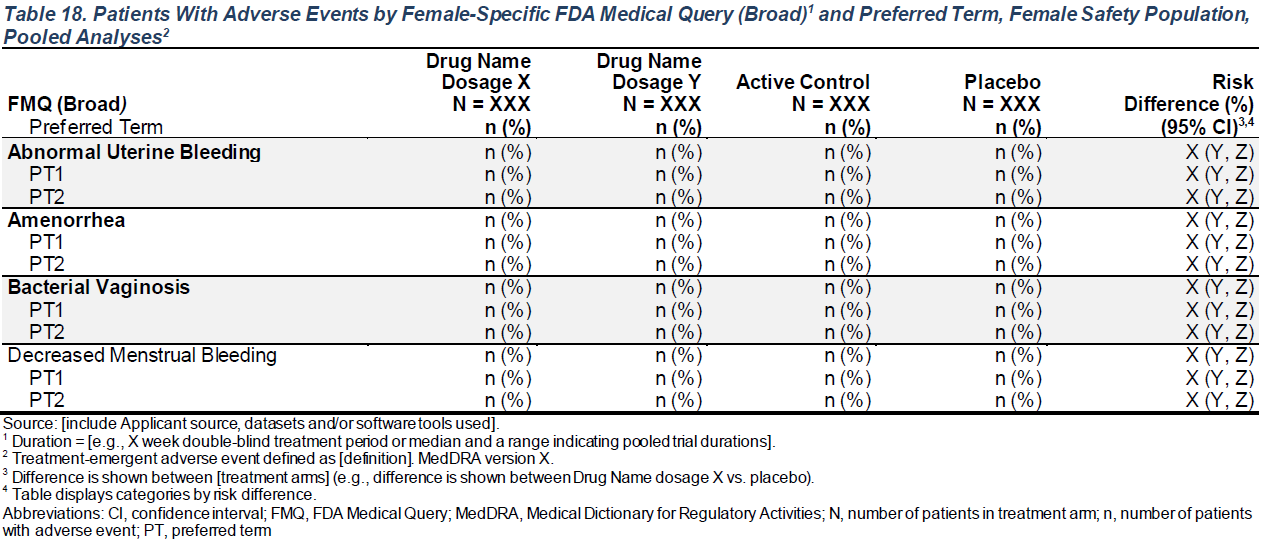{fig-align="center"}## rtables Table```{r tbl, message=FALSE, warning=FALSE}# Load Libraries & Datalibrary(cardinal)adsl <- random.cdisc.data::cadsladae <- random.cdisc.data::cadae# Pre-Processing - Ensure required variables FMQ01SC and FMQ01NAM exist in adaeset.seed(1)adae <- dplyr::rename(adae, FMQ01SC = SMQ01SC, FMQ01NAM = SMQ01NAM)levels(adae$FMQ01SC) <- c("BROAD", "NARROW")adae$FMQ01SC[is.na(adae$FMQ01SC)] <- "NARROW"adae$FMQ01NAM <- factor(adae$FMQ01NAM, levels = c( unique(adae$FMQ01NAM), "Abnormal Uterine Bleeding", "Amenorrhea", "Bacterial Vaginosis", "Decreased Menstrual Bleeding"))adae$FMQ01NAM[adae$SEX == "F"] <- as.factor( sample(c( "Abnormal Uterine Bleeding", "Amenorrhea", "Bacterial Vaginosis", "Decreased Menstrual Bleeding" ), sum(adae$SEX == "F"), replace = TRUE))# Output Tablerisk_diff <- list(arm_x = "B: Placebo", arm_y = "A: Drug X")make_table_18(adae = adae, alt_counts_df = adsl, risk_diff = risk_diff)```## rtables Table Setup```{r tbl, eval=FALSE, echo=TRUE}```## Function Details### `make_table_18()`------------------------------------------------------------------------Required variables:- **`adae`**: `adae` must contain `USUBJID`, `SEX`, and the variables specified by `arm_var`, `saffl_var`, `pref_var`, `fmqsc_var` and `fmqnam_var`.- **`alt_counts_df`** (if specified): `USUBJID` and the variables specified by `arm_var` and `saffl_var`.+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| **Argument** | **Description** | **Default** |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `adae` | (`data.frame`) Dataset (typically ADAE) required to build table. | *No default* |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `alt_counts_df` | (`character`) Alternative dataset used only to calculate column counts. | `NULL` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `show_colcounts` | (`flag`) Whether column counts should be printed. | `TRUE` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `arm_var` | (`character`) Arm variable used to split table into columns. | `"ARM"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `saffl_var` | (`character`) Flag variable used to indicate inclusion in safety population. | `"SAFFL"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `pref_var` | (`character`) Preferred term variable to use in table. | `"AEDECOD"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `sex_scope` | (`character`) Level of `SEX` to output in table. | `"F"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `fmq_scope` | (`character`) FMQ scope value to use in table. | `"BROAD"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `fmqsc_var` | (`character`) FMQ scope variable to use in table. | `"FMQ01SC"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `fmqnam_var` | (`character`) FMQ reference name variable to use in table. | `"FMQ01NAM"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `lbl_overall` | (`character`) If specified, an overall column will be added to the table with the given value as the column label. | `NULL` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `risk_diff` | (named `list`) List of settings to apply to add a risk difference column to the table. See [`tern::add_riskdiff()`](https://insightsengineering.github.io/tern/main/reference/add_riskdiff.html) for more details. List should contain the following elements: | `NULL` || | | || | - `arm_x`: (required) the name of reference arm. | || | | || | - `arm_y`: (required) the name of the arm to compare to the reference arm. | || | | || | - `col_label`: (optional) label to use for the risk difference column. Defaults to `"Risk Difference (%) (95% CI)"`. | || | | || | - `pct`: (optional) whether the output should be returned as percentages. Defaults to `TRUE`. | |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `prune_0` | (`flag`) Whether all-zero rows should be removed from the table. | `FALSE` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `na_level` | (`character`) String to represent missing values. | `"<Missing>"` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+| `annotations` | (named `list` of `character`) List of annotations to add to the table. Valid annotation types are `title`, `subtitles`, `main_footer`, and `prov_footer`. Each name-value pair should use the annotation type as name and the desired string as value. | `NULL` |+------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+---------------+Source code for this function is available [here](https://github.com/pharmaverse/cardinal/blob/main/R/fda-table_18.R).:::