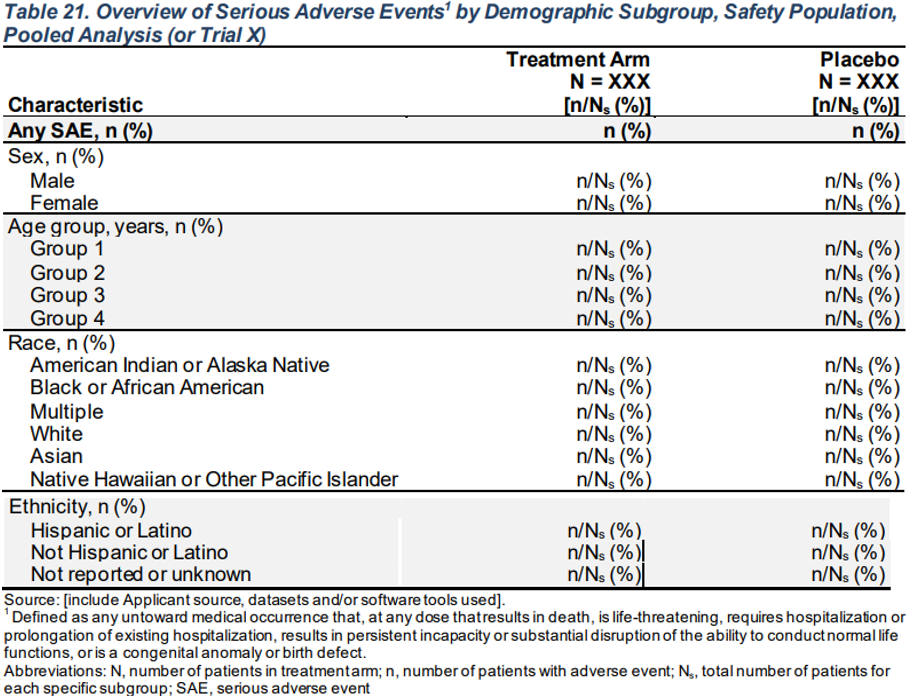
A: Drug X B: Placebo C: Combination
Characteristic (N=134) (N=134) (N=132)
————————————————————————————————————————————————————————————————————————————————————————
Any SAE 104 (77.6%) 101 (75.4%) 99 (75.0%)
SEX
F 61 (77.2%) 64 (78%) 50 (71.4%)
M 43 (78.2%) 37 (71.2%) 49 (79%)
Age Group, years
>=17 to <65 104 (77.6%) 101 (75.4%) 98 (74.8%)
>=65 0 0 1 (100%)
RACE
ASIAN 52 (76.5%) 46 (68.7%) 51 (69.9%)
BLACK OR AFRICAN AMERICAN 23 (74.2%) 23 (82.1%) 26 (81.2%)
WHITE 22 (81.5%) 22 (84.6%) 17 (81%)
AMERICAN INDIAN OR ALASKA NATIVE 7 (87.5%) 8 (72.7%) 5 (83.3%)
MULTIPLE 0 1 (100%) 0
NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER 0 1 (100%) 0
OTHER 0 0 0
UNKNOWN 0 0 0
ETHNIC
HISPANIC OR LATINO 11 (73.3%) 13 (72.2%) 11 (73.3%)
NOT HISPANIC OR LATINO 81 (77.9%) 80 (77.7%) 78 (77.2%)
NOT REPORTED 6 (100%) 7 (70%) 7 (63.6%)
UNKNOWN 6 (66.7%) 1 (33.3%) 3 (60%) FDA Table 21
Overview of Serious Adverse Events1 by Demographic Subgroup, Safety Population, Pooled Analysis (or Trial X)
# Load Libraries & Data
library(dplyr)
library(cardinal)
adsl <- random.cdisc.data::cadsl %>%
mutate(AGEGR1 = as.factor(case_when(
AGE >= 17 & AGE < 65 ~ ">=17 to <65",
AGE >= 65 ~ ">=65",
AGE >= 65 & AGE < 75 ~ ">=65 to <75",
AGE >= 75 ~ ">=75"
)) %>% formatters::with_label("Age Group, years")) %>%
formatters::var_relabel(
AGE = "Age, years"
)
adae <- random.cdisc.data::cadae
adae$ASER <- adae$AESER
df <- left_join(adsl, adae, by = intersect(names(adsl), names(adae)))
# Output Table
make_table_21(df = df, alt_counts_df = adsl, denom = "N_s")make_table_21()
Required variables:
-
df:USUBJID,ASER, and the variables specified byarm_varandsaffl_var. -
alt_counts_df(if specified):USUBJIDand the variables specified byarm_varandsaffl_var.
make_table_21() inherits from cardinal::a_count_occurrences_ser_ae() function.
| Argument | Description | Default |
|---|---|---|
df |
(data.frame) Dataset (typically ADSL) required to build table. |
No default |
alt_counts_df |
(character) Alternative dataset used only to calculate column counts. |
NULL |
show_colcounts |
(flag) Whether column counts should be printed. |
TRUE |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
vars |
(character) Variables from df to include in the table. |
c("SEX", "AGE", "AGEGR1", "RACE", "ETHNIC", "COUNTRY") |
denom |
(character) Denominator choice (N_col to get the total column, n to have the total number of “Any SAE” in the column, or N_s to have the total number of participants of the category in the column as denominator). |
c("N_s", "N_col", "n") |
lbl_vars |
(character) Labels corresponding to variables in vars to print in the table. Labels should be ordered according to the order of variables in vars. |
formatters::var_labels(df, fill = TRUE)[vars] |
lbl_overall |
(character) If specified, an overall column will be added to the table with the given value as the column label. |
NULL |
prune_0 |
(flag) Whether all-zero rows should be removed from the table. |
TRUE |
annotations |
(named list of character) List of annotations to add to the table. Valid annotation types are title, subtitles, main_footer, and prov_footer. Each name-value pair should use the annotation type as name and the desired string as value. |
NULL |
Source code for this function is available here.