# Load Libraries & Data
library(cardinal)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Pre-Processing - Ensure required variable DCSREAS exist in adae
adae$DCSREAS[is.na(adae$DCSREAS)] <- "ADVERSE EVENT"
# Select Preferred Term Variable
pref_var <- "AEDECOD"
# Output Table
make_table_12(df = adae, denominator = adsl, return_ard = FALSE, pref_var = pref_var)FDA Table 12
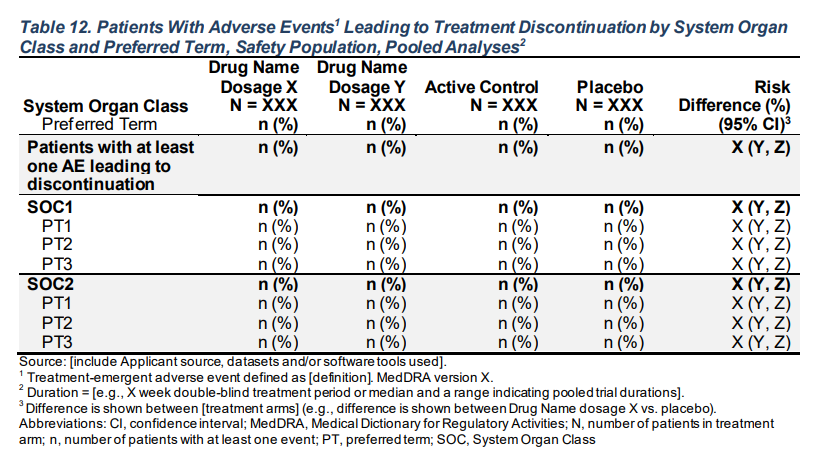
Patients With Adverse Events Leading to Treatment Discontinuation by System Organ Class and Preferred Term, Safety Population, Pooled Analyses
gtsummary Table Setup
|
Body System or Organ Class Dictionary-Derived Term |
A: Drug X N = 1341 |
B: Placebo N = 1341 |
C: Combination N = 1321 |
|---|---|---|---|
| Number of patients with event | 86 (64%) | 93 (69%) | 90 (68%) |
| cl A.1 | 53 (40%) | 55 (41%) | 69 (52%) |
| dcd A.1.1.1.1 | 37 (28%) | 37 (28%) | 50 (38%) |
| dcd A.1.1.1.2 | 33 (25%) | 31 (23%) | 40 (30%) |
| cl B.1 | 35 (26%) | 41 (31%) | 36 (27%) |
| dcd B.1.1.1.1 | 35 (26%) | 41 (31%) | 36 (27%) |
| cl B.2 | 55 (41%) | 56 (42%) | 65 (49%) |
| dcd B.2.1.2.1 | 36 (27%) | 33 (25%) | 35 (27%) |
| dcd B.2.2.3.1 | 34 (25%) | 41 (31%) | 41 (31%) |
| cl C.1 | 28 (21%) | 36 (27%) | 32 (24%) |
| dcd C.1.1.1.3 | 28 (21%) | 36 (27%) | 32 (24%) |
| cl C.2 | 26 (19%) | 39 (29%) | 43 (33%) |
| dcd C.2.1.2.1 | 26 (19%) | 39 (29%) | 43 (33%) |
| cl D.1 | 51 (38%) | 51 (38%) | 60 (45%) |
| dcd D.1.1.1.1 | 29 (22%) | 31 (23%) | 37 (28%) |
| dcd D.1.1.4.2 | 33 (25%) | 34 (25%) | 39 (30%) |
| cl D.2 | 35 (26%) | 46 (34%) | 44 (33%) |
| dcd D.2.1.5.3 | 35 (26%) | 46 (34%) | 44 (33%) |
| 1 n (%) | |||
Function Details
make_table_12()
Required variables:
-
df:USUBJID,AEBODSYS,DCSREAS, and the variables specified bypref_var,arm_var, andsaffl_var. -
alt_counts_df(if specified):USUBJIDand the variables specified byarm_varandsaffl_var.
| Argument | Description | Default |
df |
(data.frame) Dataset (typically ADSL) required to build table. |
No default |
return_ard |
(flag) Whether an ARD should be returned. |
TRUE |
denominator |
(character) Alternative dataset used only to calculate column counts. |
NULL |
id_var |
(character) Identifier variable used to count the participants within each flag. |
"USUBJID" |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
pref_var |
(character) Name of the preferred term variable from adae to include in the table. |
"AEDECOD" |
ARD Setup
# Load Libraries & Data
library(cardinal)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Pre-Processing - Ensure required variable DCSREAS exist in adae
adae$DCSREAS[is.na(adae$DCSREAS)] <- "ADVERSE EVENT"
# Select Preferred Term Variable
pref_var <- "AEDECOD"
# Create Table & ARD
result <- make_table_12(df = adae, denominator = adsl, pref_var = pref_var)
# Output ARD
result$ard{cards} data frame: 90 x 13 group1 group1_level group2 group2_level variable variable_level context stat_name stat_label stat fmt_fn warning error
1 ARM A: Drug X AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… n n 51 0
2 ARM A: Drug X AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… N N 134 0
3 ARM A: Drug X AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… p % 0.381 <fn>
4 ARM A: Drug X AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… n n 45 0
5 ARM A: Drug X AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… N N 134 0
6 ARM A: Drug X AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… p % 0.336 <fn>
7 ARM A: Drug X AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… n n 42 0
8 ARM A: Drug X AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… N N 134 0
9 ARM A: Drug X AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… p % 0.313 <fn>
10 ARM A: Drug X AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… n n 48 0
11 ARM A: Drug X AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… N N 134 0
12 ARM A: Drug X AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… p % 0.358 <fn>
13 ARM A: Drug X AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… n n 47 0
14 ARM A: Drug X AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… N N 134 0
15 ARM A: Drug X AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… p % 0.351 <fn>
16 ARM A: Drug X AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… n n 36 0
17 ARM A: Drug X AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… N N 134 0
18 ARM A: Drug X AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… p % 0.269 <fn>
19 ARM A: Drug X AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… n n 35 0
20 ARM A: Drug X AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… N N 134 0
21 ARM A: Drug X AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… p % 0.261 <fn>
22 ARM A: Drug X AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… n n 31 0
23 ARM A: Drug X AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… N N 134 0
24 ARM A: Drug X AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… p % 0.231 <fn>
25 ARM A: Drug X AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… n n 46 0
26 ARM A: Drug X AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… N N 134 0
27 ARM A: Drug X AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… p % 0.343 <fn>
28 ARM A: Drug X AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… n n 44 0
29 ARM A: Drug X AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… N N 134 0
30 ARM A: Drug X AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… p % 0.328 <fn>
31 ARM B: Place… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… n n 52 0
32 ARM B: Place… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… N N 134 0
33 ARM B: Place… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… p % 0.388 <fn>
34 ARM B: Place… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… n n 40 0
35 ARM B: Place… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… N N 134 0
36 ARM B: Place… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… p % 0.299 <fn>
37 ARM B: Place… AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… n n 51 0
38 ARM B: Place… AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… N N 134 0
39 ARM B: Place… AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… p % 0.381 <fn>
40 ARM B: Place… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… n n 44 0
41 ARM B: Place… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… N N 134 0
42 ARM B: Place… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… p % 0.328 <fn>
43 ARM B: Place… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… n n 57 0
44 ARM B: Place… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… N N 134 0
45 ARM B: Place… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… p % 0.425 <fn>
46 ARM B: Place… AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… n n 47 0
47 ARM B: Place… AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… N N 134 0
48 ARM B: Place… AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… p % 0.351 <fn>
49 ARM B: Place… AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… n n 43 0
50 ARM B: Place… AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… N N 134 0
51 ARM B: Place… AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… p % 0.321 <fn>
52 ARM B: Place… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… n n 40 0
53 ARM B: Place… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… N N 134 0
54 ARM B: Place… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… p % 0.299 <fn>
55 ARM B: Place… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… n n 46 0
56 ARM B: Place… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… N N 134 0
57 ARM B: Place… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… p % 0.343 <fn>
58 ARM B: Place… AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… n n 58 0
59 ARM B: Place… AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… N N 134 0
60 ARM B: Place… AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… p % 0.433 <fn>
61 ARM C: Combi… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… n n 73 0
62 ARM C: Combi… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… N N 132 0
63 ARM C: Combi… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… p % 0.553 <fn>
64 ARM C: Combi… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… n n 56 0
65 ARM C: Combi… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… N N 132 0
66 ARM C: Combi… AEBODSYS cl A.1 AEDECOD dcd A.1.… hierarch… p % 0.424 <fn>
67 ARM C: Combi… AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… n n 52 0
68 ARM C: Combi… AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… N N 132 0
69 ARM C: Combi… AEBODSYS cl B.1 AEDECOD dcd B.1.… hierarch… p % 0.394 <fn>
70 ARM C: Combi… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… n n 44 0
71 ARM C: Combi… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… N N 132 0
72 ARM C: Combi… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… p % 0.333 <fn>
73 ARM C: Combi… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… n n 62 0
74 ARM C: Combi… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… N N 132 0
75 ARM C: Combi… AEBODSYS cl B.2 AEDECOD dcd B.2.… hierarch… p % 0.47 <fn>
76 ARM C: Combi… AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… n n 53 0
77 ARM C: Combi… AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… N N 132 0
78 ARM C: Combi… AEBODSYS cl C.1 AEDECOD dcd C.1.… hierarch… p % 0.402 <fn>
79 ARM C: Combi… AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… n n 49 0
80 ARM C: Combi… AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… N N 132 0
81 ARM C: Combi… AEBODSYS cl C.2 AEDECOD dcd C.2.… hierarch… p % 0.371 <fn>
82 ARM C: Combi… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… n n 51 0
83 ARM C: Combi… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… N N 132 0
84 ARM C: Combi… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… p % 0.386 <fn>
85 ARM C: Combi… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… n n 52 0
86 ARM C: Combi… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… N N 132 0
87 ARM C: Combi… AEBODSYS cl D.1 AEDECOD dcd D.1.… hierarch… p % 0.394 <fn>
88 ARM C: Combi… AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… n n 55 0
89 ARM C: Combi… AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… N N 132 0
90 ARM C: Combi… AEBODSYS cl D.2 AEDECOD dcd D.2.… hierarch… p % 0.417 <fn> rtables Table Setup
# Load Libraries & Data
library(cardinal)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Pre-Processing - Ensure required variable DCSREAS exist in adae
adae$DCSREAS[is.na(adae$DCSREAS)] <- "ADVERSE EVENT"
# Select Preferred Term Variable
pref_var <- "AEDECOD"
# Output Table
risk_diff <- list(arm_x = "B: Placebo", arm_y = "A: Drug X") # optional
make_table_12_rtables(df = adae, alt_counts_df = adsl, pref_var = pref_var, risk_diff = risk_diff)Body System or Organ Class A: Drug X B: Placebo C: Combination Risk Difference (%) (95% CI)
Dictionary-Derived Term (N=134) (N=134) (N=132) (N=268)
——————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
Patients with at least one AE leading to discontinuation 86 (64.2%) 93 (69.4%) 90 (68.2%) 5.2 (-6.0 - 16.5)
cl A.1 53 (39.6%) 55 (41.0%) 69 (52.3%) 1.5 (-10.3 - 13.2)
dcd A.1.1.1.1 37 (27.6%) 37 (27.6%) 50 (37.9%) 0.0 (-10.7 - 10.7)
dcd A.1.1.1.2 33 (24.6%) 31 (23.1%) 40 (30.3%) -1.5 (-11.7 - 8.7)
cl B.1 35 (26.1%) 41 (30.6%) 36 (27.3%) 4.5 (-6.3 - 15.3)
dcd B.1.1.1.1 35 (26.1%) 41 (30.6%) 36 (27.3%) 4.5 (-6.3 - 15.3)
cl B.2 55 (41.0%) 56 (41.8%) 65 (49.2%) 0.7 (-11.0 - 12.5)
dcd B.2.1.2.1 36 (26.9%) 33 (24.6%) 35 (26.5%) -2.2 (-12.7 - 8.2)
dcd B.2.2.3.1 34 (25.4%) 41 (30.6%) 41 (31.1%) 5.2 (-5.5 - 16.0)
cl C.1 28 (20.9%) 36 (26.9%) 32 (24.2%) 6.0 (-4.2 - 16.2)
dcd C.1.1.1.3 28 (20.9%) 36 (26.9%) 32 (24.2%) 6.0 (-4.2 - 16.2)
cl C.2 26 (19.4%) 39 (29.1%) 43 (32.6%) 9.7 (-0.5 - 19.9)
dcd C.2.1.2.1 26 (19.4%) 39 (29.1%) 43 (32.6%) 9.7 (-0.5 - 19.9)
cl D.1 51 (38.1%) 51 (38.1%) 60 (45.5%) 0.0 (-11.6 - 11.6)
dcd D.1.1.1.1 29 (21.6%) 31 (23.1%) 37 (28.0%) 1.5 (-8.5 - 11.5)
dcd D.1.1.4.2 33 (24.6%) 34 (25.4%) 39 (29.5%) 0.7 (-9.6 - 11.1)
cl D.2 35 (26.1%) 46 (34.3%) 44 (33.3%) 8.2 (-2.7 - 19.2)
dcd D.2.1.5.3 35 (26.1%) 46 (34.3%) 44 (33.3%) 8.2 (-2.7 - 19.2) Function Details
make_table_12_rtables()
Required variables:
-
df:USUBJID,AEBODSYS,DCSREAS, and the variables specified bypref_var,arm_var, andsaffl_var. -
alt_counts_df(if specified):USUBJIDand the variables specified byarm_varandsaffl_var.
| Argument | Description | Default |
adae |
(data.frame) Dataset (typically ADAE) required to build table. |
No default |
alt_counts_df |
(character) Alternative dataset used only to calculate column counts. |
NULL |
show_colcounts |
(flag) Whether column counts should be printed. |
TRUE |
arm_var |
(character) Arm variable used to split table into columns. |
"ARM" |
saffl_var |
(character) Flag variable used to indicate inclusion in safety population. |
"SAFFL" |
pref_var |
(character) Preferred term variable from adae to include in the table. |
"AEDECOD" |
lbl_overall |
(character) If specified, an overall column will be added to the table with the given value as the column label. |
NULL |
risk_diff |
(named
|
NULL |
prune_0 |
(flag) Whether all-zero rows should be removed from the table. |
FALSE |
annotations |
(named list of character) List of annotations to add to the table. Valid annotation types are title, subtitles, main_footer, and prov_footer. Each name-value pair should use the annotation type as name and the desired string as value. |
NULL |